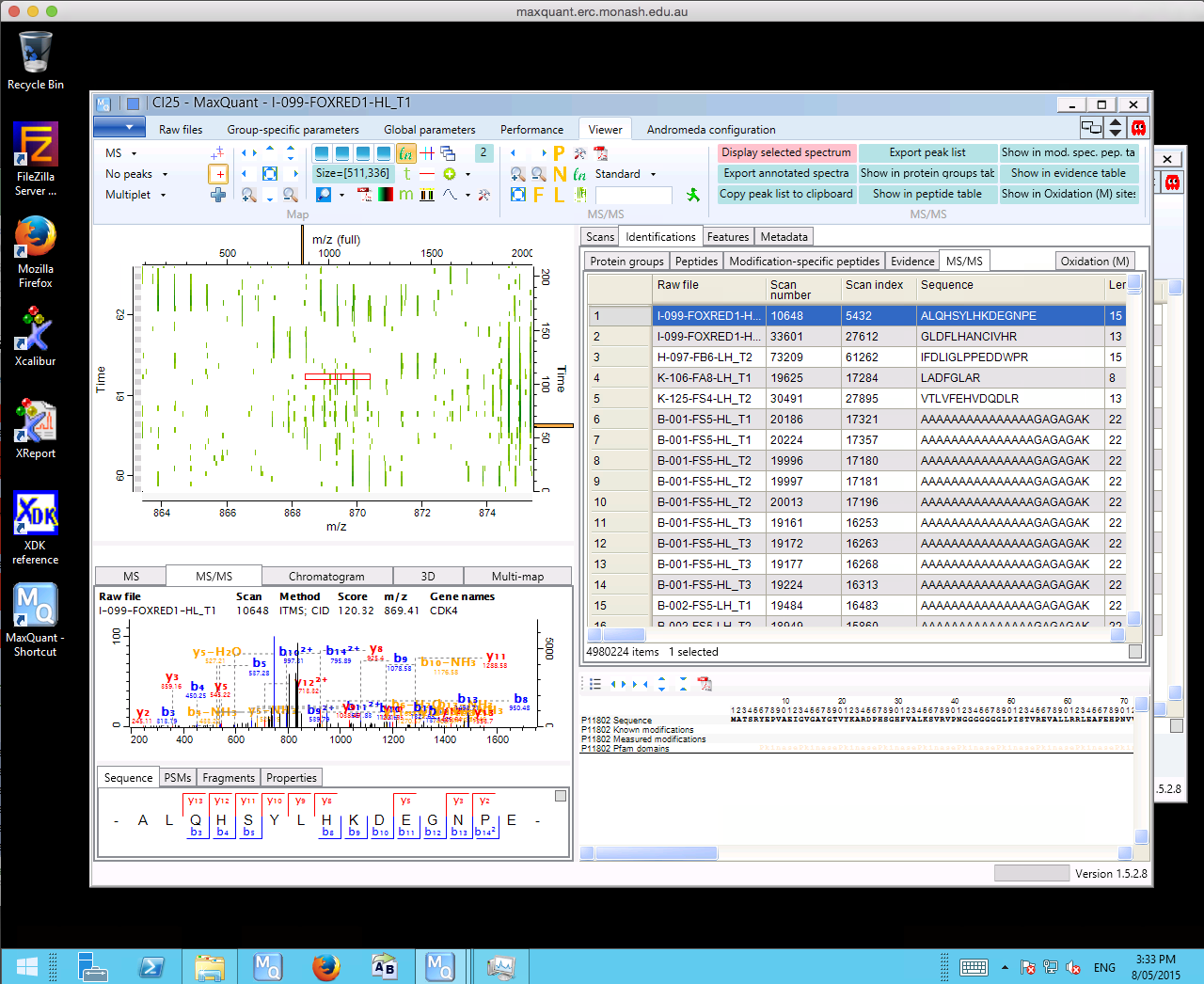
David Stroud, NHMRC Doherty Fellow and member of the Ryan Lab from the Department of Biochemistry and Molecular Biology, Monash University does proteomics research and uses the MaxQuant quantitative proteomics software as part of his analysis workflows. MaxQuant is designed for processing high-resolution Mass Spectrometry data and is freely available on the Microsoft Windows platform. Step one in the workflow is to do sample analyses using Liquid chromatography-mass spectrometry (LC-MS) on a Thermo Orbitrap Mass-spectrometer. This step produces raw files containing spectra that represent thousands of peptides. The resulting raw files are then loaded into MaxQuant to perform searches where spectra are compared against known list of peptides. A quantification step is then performed enabling peptide abundance to be compared across samples. Once this process is completed, the resulting tab delimited files are captured for downstream analysis.
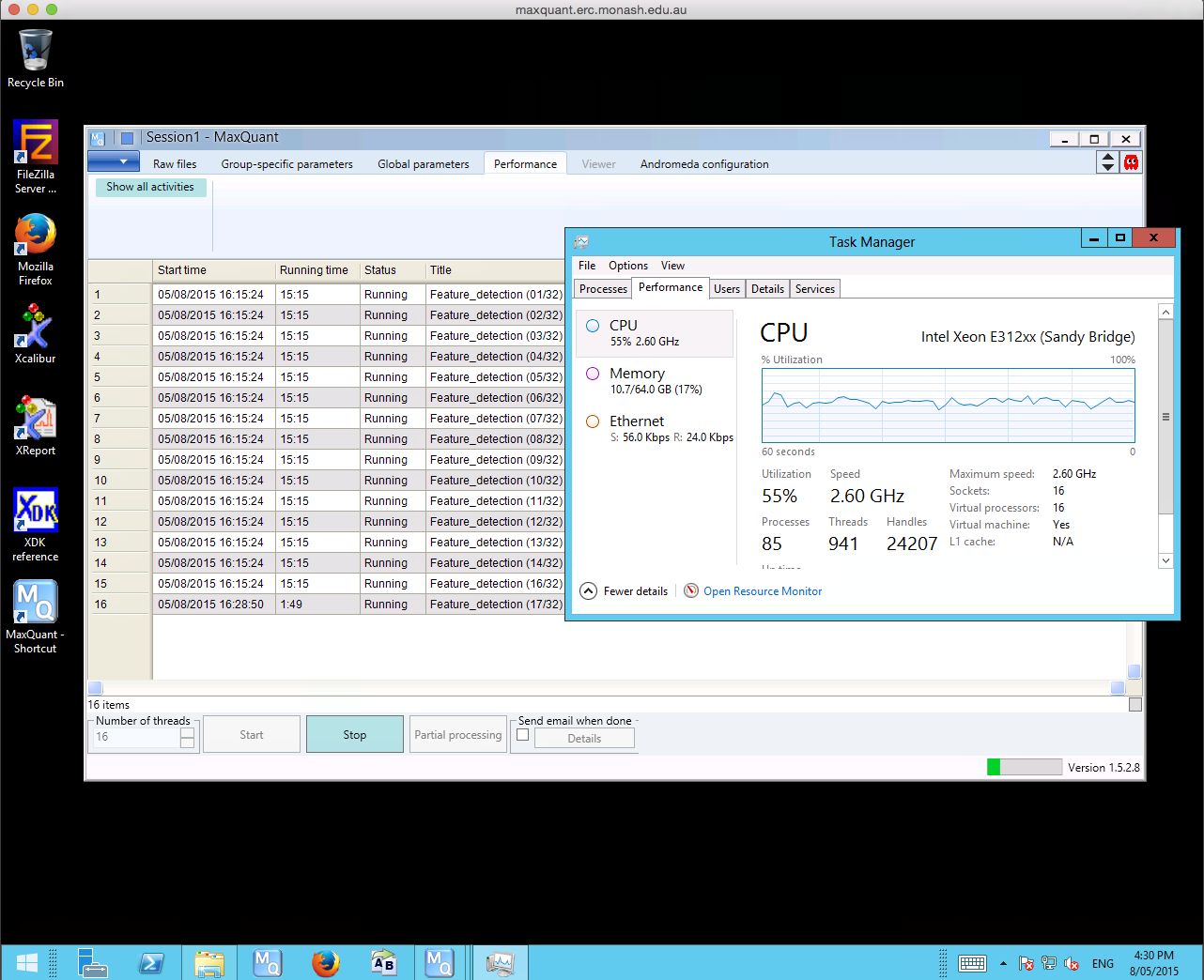
MaxQuant searches are both CPU and IO intensive tasks. A typical search takes 24 to 48 hours, and in some cases up to a week, depending on the size of the raw files being processed. David has been running his workflow on his own machine with 8 cores, 16 gigabytes of memory (RAM) and a solid state drive (SSD) for storage where a standard search takes 2 to 3 weeks to complete. Performing large MaxQuant searches on the local machine became a struggle, and David needed a bigger machine with a desktop environment to scale up his analysis workflow. The R@CMon team assisted David in deploying the MaxQuant software on the Monash node of the NeCTAR Research Cloud with an m1.xxlarge instance, spawned using the Monash-licensed Windows Server 2012 image. MaxQuant searches on the NeCTAR instance shows a 3-4x speed-up compared to the local machine, what takes several weeks on the local machine now just takes several days on the NeCTAR instance.
Maxquant search of Thermo RAW files.
The R@CMon team are currently working with David to explore further scaling options. The high-memory and PCIe SSD-enabled specialist kit on R@CMon Phase 2 can be exploited by MaxQuant for bursting IO intensive activities during searches. More on this coming soon!