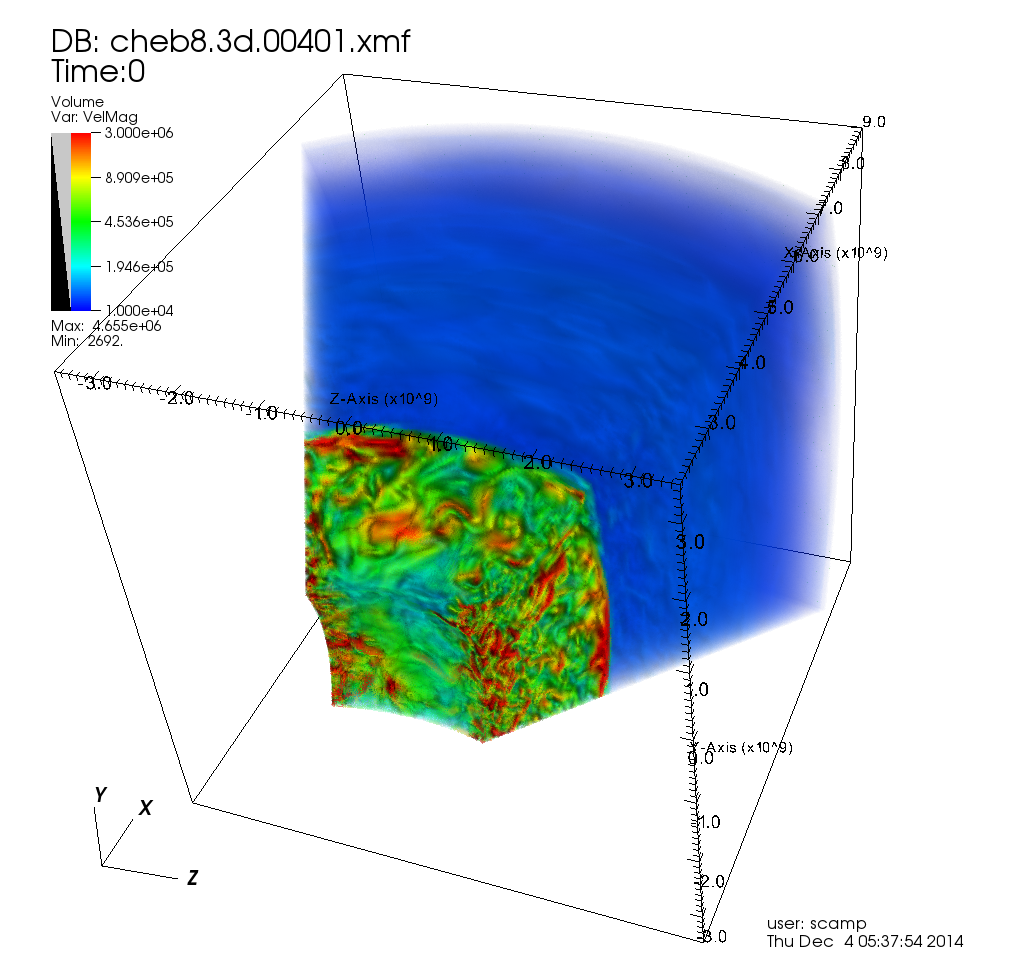
Monash is home to the Multi-modal Australian ScienceS Imaging and Visualisation Environment (MASSIVE), a national facility for the imaging and characterisation community. An important and rather novel feature of the MASSIVE compute cluster is the interactive desktop visualisation environment available to assist users in the characterisation process. The MASSIVE desktop environment provided part of the inspiration for the Characterisation Virtual Laboratory (CVL), a NeCTAR VL project combining specialist software visualisation and rendering tools from a variety of disciplines and making them available on and through the NeCTAR research cloud.
The recently released monash-02 zone of the NeCTAR cloud provides enhanced capability to the CVL, bringing a critical mass of GPU accelerated cloud instances. monash-02 includes ten GPU capable hypervisors, currently able to provide up to thirty GPU accelerated instances via direct PCI passthrough. Most of these are NVIDIA GRID K2 GPUs (CUDA 3.0 capable), though we also have one K1. Special thanks to NVIDIA for providing us with a couple of seed units to get this going and supplement our capacity! After consultation with various users we created the following set of flavors/instance-types for these GPUs:
| Flavor name | #vcores | RAM (MB) | /dev/vda (GB) | /dev/vdb (GB) |
|---|---|---|---|---|
| mon.r2.5.gpu-k2 | 1 | 5400 | 30 | N/A |
| mon.r2.10.gpu-k2 | 2 | 10800 | 30 | 40 |
| mon.r2.21.gpu-k2 | 4 | 21700 | 30 | 160 |
| mon.r2.63.gpu-k2 | 12 | 65000 | 30 | 320 |
| mon.r2.5.gpu-k1 | 1 | 5400 | 30 | N/A |
| mon.r2.10.gpu-k1 | 2 | 10800 | 30 | 40 |
| mon.r2.21.gpu-k1 | 4 | 21700 | 30 | 160 |
| mon.r2.63.gpu-k1 | 12 | 65000 | 30 | 320 |
R@CMon has so far dedicated two of these GPU nodes to the CVL, and this is our preferred method for use of this equipment, as the CVL provides a managed environment and queuing system for access (regular plain IaaS usage is available where needed). There were some initial hiccups getting the CVL’s base CentOS 6.6 image working with NVIDIA drivers on these nodes, solved by moving to a newer kernel, and some performance tuning tasks still remain. However, the CVL has now been updated to make use of the new GPU flavors on monash-02, as demonstrated in the following video…
GPU-accelerated Chimera application running on the CVL, showing the structure of human follicle-stimulating hormone (FSH) and its receptor.
If you’re interested in using GPGPUs on the cloud please contact the R@CMon team or Monash eResearch Centre.