Glycosylation is an ubiquitous type of protein post-translational modification (PTM) in eukaryotic cells, which plays vital roles in various biological processes such as cellular communication, ligand recognition and subcellular recognition. It is estimated that greater than 50% of the entire human proteome is glycosylated. However, it is still a significant challenge to identify glycosylation sites, which requires expensive and laborious experimental research. Thus, bioinformatics approaches that can predict the glycan occupancy at specific sequons in protein sequences would be useful for understanding and utilising this very important PTM.
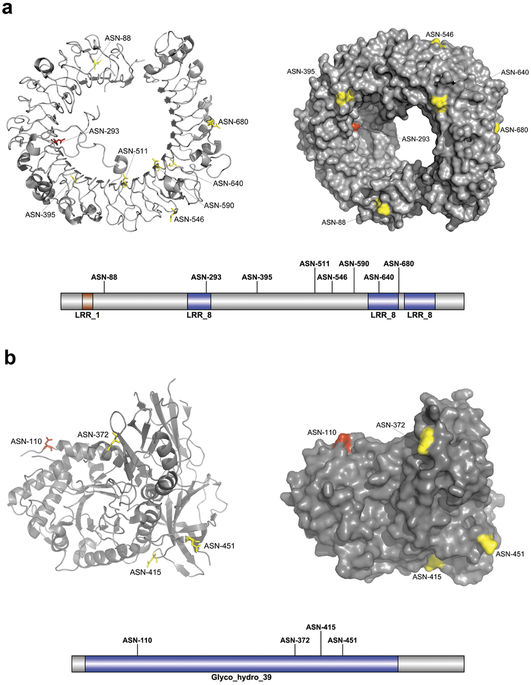
Predicted N-linked glycosylation sites from two case-study proteins using GlycoMine-Struct
Dr. Jiangning Song from the Department of Biochemistry and Molecular Biology at Monash University and his collaborators have designed and developed a bioinformatics tool – GlycoMine-Struct for predicting glycosylation sites. GlycoMine-Struct is a comprehensive tool for the systematic in-silico identification of N-linked and O-linked glycosylation sites in the human proteome. Through R@CMon, a dedicated cloud project with computational and storage resources has been provisioned to develop and host the GlycoMine-Struct tool. The flexible and scalable R@CMon-powered development environment enabled rapid prototyping, testing and re-deployment of the tool.
GlycoMine-Struct Main Page (http://glycomine.erc.monash.edu/Lab/GlycoMine_Struct/index.jsp#Introduction)
GlycoMine-Struct is now a publicly accessible web service, available to the wider research community. Users can now easily submit protein structure input files in PDB (Protein Data Bank) format to perform sites prediction on GlycoMine-Struct. Since it went public, GlycoMine-Struct has been accessed and used by thousands of local and international users, and still growing. A scientific reports paper has been published, highlighting the collaborative work done to develop GlycoMine-Struct, as an essential bioinformatics tool for improving the prediction of human glycosylation sites. The R@CMon team is actively supporting the GlycoMine-Struct project as it continues to serve the research community and develop performance improvements.