| Glossary Terms: DOCA = Data Center-on-a-Chip Architecture DPU = Data Processing Unit NIC = Network Interface Card OVS = Open Virtual Switch (also known as Open vSwitch) SF = Scalable Function VF = Virtual Function VM = Virtual Machine |
Motivation
In part one of this series, we showed how to set up an NVIDIA BlueField-2 DPU with the aim of simply using it as a NIC (https://rcblog.erc.monash.edu.au/blog/2022/02/how-do-i-use-a-dpu-as-nic/). Today we’re going to delve a bit deeper and offload network layer encryption to the DPUs so that VMs running on the Host can use more of their allocated resources while still communicating securely.
Background
Servers and processors are becoming faster and more powerful, but at the same time the requirement for greater network security is increasing and end-to-end network encryption is mandatory in some of the more sensitive data research workloads. As the data throughput on the CPU is pushed to the limits, processors start to steal compute cycles for the network to be encrypted away from computing tasks for research work.
Our aim is to try to mitigate part of this issue by offloading the work of encryption and decryption of traffic from the processors on the hypervisor(host) to the DPU and enable our researchers to work more efficiently and securely.
Set up
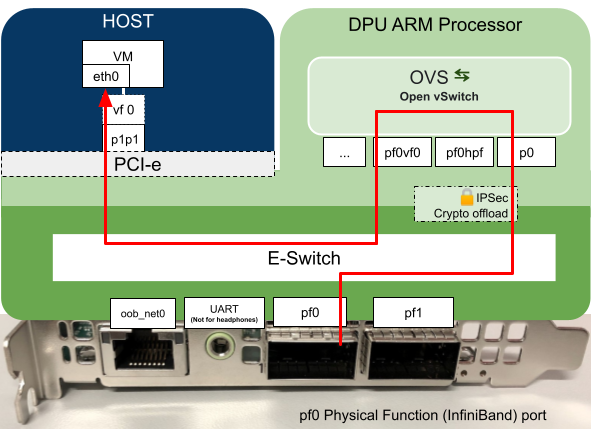
Below is a diagram of the setup we used while testing:
Figure: Diagram of the setup we are describing. Two hosts running one VM each, each host has a DPU installed in a PCI slot, the DPUs are connected directly to each other (back to back).
As we explained in our previous blog post, the DPU’s have programmable hardware for data processing, in this example the DPU has been programmed to process data coming in on port p0 which is then passed to the host which in turn is presented to the VM as the Virtual Function (vf0). The virtual function interface appears at the VM as eth1.
Setting up the DPU
Once the environment is set up as above we can move on to configure Open vSwitch and strongSwan encryption software on the DPU(yes that is the correct capitalization stylization)
To set up the OVS bridges and strongSwan (5.9.0bf) IPSec tunnel we used NVIDIA’s example from this website
https://docs.nvidia.com/doca/sdk/east-west-overlay-encryption/index.html
The change we needed to make was to add the interface pf0vf0 to the OVS Bridge vxlan-br0 on both DPUs to allow the VM’s to communicate with each other.
Example of OVS settings from one of the DPUs
| ubuntu@localhost:~$ sudo ovs-vsctl show 352ec404-2519-4751-9a1b-3fd33780543c Bridge vxlan-br0 Port pf0vf0 Interface pf0vf0 Port vxlan-br0 Interface vxlan-br0 type: internal Port vxlan11 Interface vxlan11 type: vxlan options: {dst_port=”4789″, key=”100″, local_ip=”192.168.50.1″, remote_ip=”192.168.50.2″} Port pf0hpf Interface pf0hpf ovs_version: “2.15.1-d246dab” |
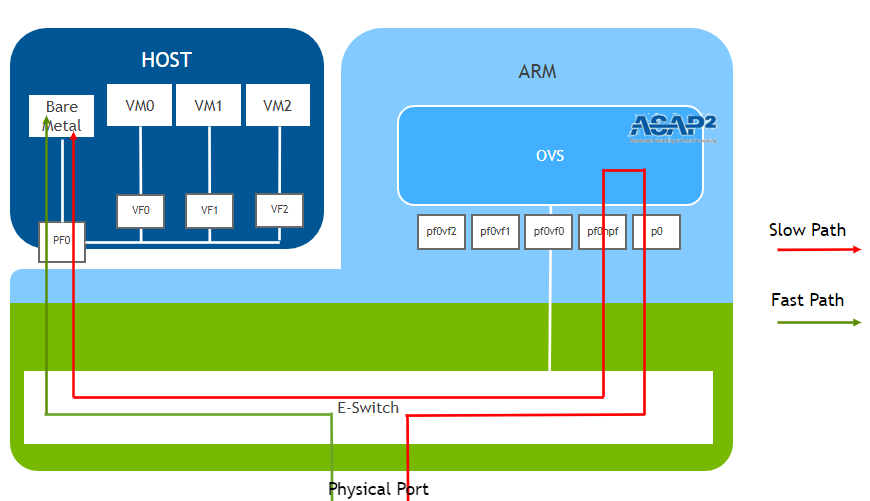
With the above OVS configuration, the logical flow of packets will be as below.
Here is a diagram of the “Path” defined by the above configuration:
Diagram of the logical setup:
Figure: Logical flow of packets through the DPU to the VM.
Our current setup uses the Slow Path
The diagram was referenced on NVIDIA’s original from (https://docs.nvidia.com/doca/sdk/l4-ovs-firewall/index.html)
Setting IPSec Full Offload Using strongSwan
strongSwan configures IPSec HW full offload using a new value added to its configuration file.
By default two files are created in /etc/swanctl/conf.d when flashing the DPUs with DOCA SDK.
BFL.swanctl.conf and BFR.swanctl.conf
We only want one of these on each host. BFL on Host 16 and BFR on Host 17
We also want to make some changes to the .conf files.
On DPU 16
| cd /etc/swanctl/conf.d/ mv BFR.swanctl.conf BFR.swanctl.conf.old vi /etc/swanctl/conf.d/BFL.swanctl.conf #Note edit this file manually, copying the below output will probably result in issues cat /etc/swanctl/conf.d/BFL.swanctl.conf # LEFT: strongswan BF-2 config file connections { BFL-BFR { local_addrs = 3.3.3.2 remote_addrs = 3.3.3.3 local { auth = psk id = host2 } remote { auth = psk id = host1 } children { bf { local_ts = 3.3.3.2/24 [udp/4789] remote_ts = 3.3.3.3/24 [udp/4789] esp_proposals = aes128gcm128-x25519-esn mode = transport policies_fwd_out = yes hw_offload = full } } version = 2 mobike = no reauth_time = 0 proposals = aes128-sha256-x25519 } } secrets { ike-BF { id-host1 = host1 id-host2 = host2 secret = 0sv+NkxY9LLZvwj4qCC2o/gGrWDF2d21jL } } |
On DPU 17
| cd /etc/swanctl/conf.d/ mv BFL.swanctl.conf BFL.swanctl.conf.old vi /etc/swanctl/conf.d/BFR.swanctl.conf #Note edit this file manually, copying the below output will probably result in issues cat /etc/swanctl/conf.d/BFR.swanctl.conf # RIGHT: strongswan BF-2 config file connections { BFL-BFR { local_addrs = 3.3.3.3 remote_addrs = 3.3.3.2 local { auth = psk id = host1 } remote { auth = psk id = host2 } children { bf { local_ts = 3.3.3.3/24 [udp/4789] remote_ts = 3.3.3.2/24 [udp/4789] esp_proposals = aes128gcm128-x25519-esn mode = transport policies_fwd_out = yes hw_offload = full } } version = 2 mobike = no reauth_time = 0 proposals = aes128-sha256-x25519 } } secrets { ike-BF { id-host1 = host1 id-host2 = host2 secret = 0sv+NkxY9LLZvwj4qCC2o/gGrWDF2d21jL } } |
Note: Make sure there is a new line at the end of these files or the config may not be applied correctly.
Commands to load strongSwan configuration
| On Both DPUs systemctl stop strongswan-starter.service systemctl start strongswan-starter.service swanctl –load-all On left DPU (DPU 16) swanctl -i –child bf |
Commands to switch offloading on and off
| #To enable offloading: ovs-vsctl set Open_vSwitch . Other_config:hw-offload=true systemctl restart openvswitch-switch #To disable offloading: ovs-vsctl –no-wait set Open_vSwitch . other_config:hw-offload=false systemctl restart openvswitch-switch #Check current offloading state ovs-vsctl get Open_vSwitch . other_config:hw-offload |
Experiments and results
We can now transmit data between the VMs which will automatically be encrypted by the DPUs as the information goes over the wire. We use iperf3 to generate network traffic between the VM’s while switching on and off hardware offloading capability on the DPU.
We start with hw-offloading disabled to observe the following results
Offload Disabled
| #To disable offloading: ovs-vsctl –no-wait set Open_vSwitch . other_config:hw-offload=false systemctl restart openvswitch-switch |
With offloading turned off via OVS on the DPU, we can see a ksoftirqd process with high cpu utilisation in the DPU.
Next we turn on hw-offloading and observed the change in behaviour
Offload Enabled
| #To enable offloading: ovs-vsctl set Open_vSwitch . Other_config:hw-offload=true systemctl restart openvswitch-switch |
With hw-offload enabled we no longer see the ksoftirqd having high CPU utilisation on the DPU
And the rate of data transfer increased more than 10 times than what we were seeing with hw-offload disabled.
We can also view the offloaded flows on the DPU when hw-offload is enabled:
| ovs-appctl dpctl/dump-flows type=offloaded #Example output root@localhost:/home/ubuntu# ovs-appctl dpctl/dump-flows type=offloaded recirc_id(0),in_port(4),eth(src=7a:9a:41:5d:11:81,dst=1e:e1:b3:b7:cb:db),eth_type(0x0800),ipv4(tos=0/0x3,frag=no), packets:3987309, bytes:6229832925, used:0.710s, actions:set(tunnel(tun_id=0x64,src=192.168.50.2,dst=192.168.50.1,ttl=64,tp_dst=4789,flags(key))),3 tunnel(tun_id=0x64,src=192.168.50.1,dst=192.168.50.2,tp_dst=4789,flags(+key)),recirc_id(0),in_port(3),eth(src=1e:e1:b3:b7:cb:db,dst=7a:9a:41:5d:11:81),eth_type(0x0800),ipv4(frag=no), packets:49131, bytes:5899556, used:0.710s, actions:4 |
Which proves that the offloaded flows were indeed executed on the DPU.
The above output shows the offloaded flows and their source IP address, destination IP address, source port, destination port, protocol and the number of packets and bytes in the flow.
Conclusion
At this point we’ve set up two VMs and built an ipsec tunnel between them on the DPUs which automatically encrypts data sent over the wire.
We’ve shown a noticeable speed difference between enabling and disabling hw-offload and shown how to check the flows when they are being offloaded and the impact on performance when they are not being offloaded.
Future
As we provide IaaS (Infrastructure-as-a-Service) using OpenStack, the next step is to automate the setup of this configuration in the infrastructure using our CICD configuration management at DPUs so that the network traffic from the research workload VMs are encrypted underneath. We also consider having dedicated IPSec tunnels for the VMs from different projects running on the host so that underlay encrypted network fabric is separated for each project.
Written by Ben Boreham, Shahaan Ayyub, Swe Aung and Steve Quenette as part of a partnership between Nvidia, the Australian Research Data Commons (ARDC), and Monash University