Precision medicine and genomics hold great potential for improved detection of cancer, particularly the targeted DNA sequencing of genes that indicate the risk of developing cancer in the future. In 2015, solving this problem required a technologically complex task of combining advanced genomics analysis with extensive medical (phenotypic) health data. The research domain wasn’t there yet. It was still exploring, and here’s a part we played with Australia’s largest clinical trial.
Associate Professor Paul Lacaze is the head of the Public Health Genomics Program within the School of Public Health and Preventive Medicine, Monash University. Since 2015 this program has formed an integral part of the ASPREE study1 and ASPREE Healthy Ageing Biobank2. Their strategy was to partner with genomic sequencing facilities from across the globe, who each bought distinct expertise to the challenge of sequencing thousands of ASPREE participants for precision medicine applications. Each partnership has since been on a mission to integrate the study’s phenotypic and clinical outcome data with its novel techniques to understand the role of genetics in healthy ageing and diseases.
Hence our multiple global research collaborations needed an environment where they could discover how to join sensitive and big data such that insights could emerge. Solving such multi-disciplinary techno-social problems is the bread & butter of the digital cooperatives group within the Monash eResearch Centre (MeRC). We activated a hybrid of HPC-like and cloud resources appropriate for the active merging of sensitive clinical trial data with the targeted DNA sequencing data to solve this problem. The researchers explored a range of computational tools & techniques that were in themselves still an experiment. Learnings from engagements like this inform the processes and procedures we have today. Most pertinently, however, we took those communities and their respective organisations through the journey, and they now enjoy low barriers to generating impacts from these collaborations.
One of the research-led sequencing technologies or techniques is the “Targeted Sequencing” (Super Panel). The program collaborated with the Icahn School of Medicine at Mount Sinai to design a panel with around ~700 distinct genes that capture the following gene groups: Cancer Genes, Cardiovascular Genes, PGX Genes, ACMG 56 Genes, Resilience Genes and Maturity Onset Diabetes of the Young (MODY) Genes.
From a logistical point of view, one of the advantages of target sequencing is the smaller storage footprint compared to whole-genome sequencing (which can be ten times bigger in terms of file size). The Mount Sinai group sequenced 13,000 ASPREE samples over several months. That generated ~30TB of sequence alignment files (BAMs) and variation files (VCFs). The first task (back then) was to establish a secure transfer channel for the data to Monash for both storage and downstream analysis. Immediately Paul identified that he did not have the tool or service readily available for this task. That’s when he engaged with MeRC. The Research Cloud at Monash (R@CMon) and digital cooperatives teams within MeRC provided the solution to address the project’s data transfer, storage, and computational requirements (see Figure 1 below). We have since co-operated this infrastructure with them.
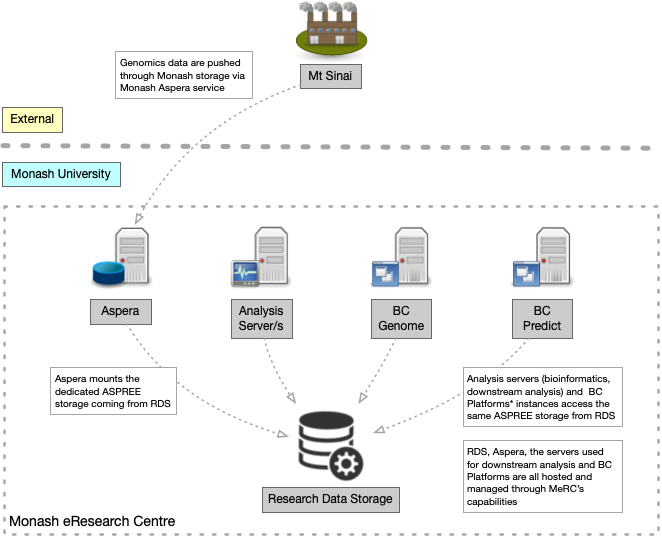
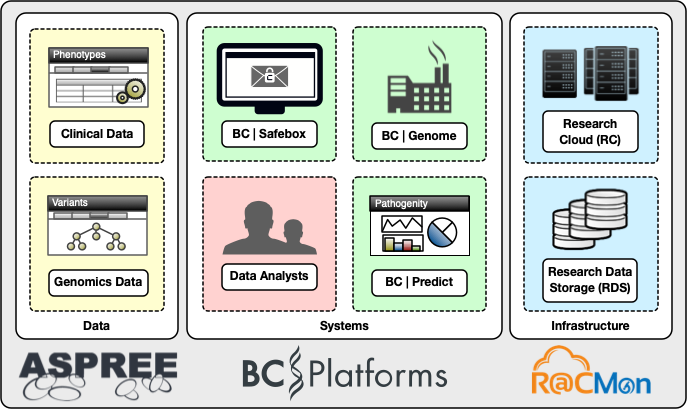
In addition to collaborating with the Public Health Genomics Program and the program collaborating with clinical genomics leaders from across the globe, we collaborated with a software vendor. Together with BC Platforms, we designed a custom-build information system that meets the requirements of both clinical and genomics data processing. The digital cooperatives team provided hosting through the Research Cloud and Research Data Storage. We configured the analysis servers deployed on the Research Cloud with bioinformatics tools for processing the genomics data. Additionally, we deployed three core commercial products from BC Platforms: BC Genome – a secure online database (data warehousing system) for storing and dynamic analysis of genotype and phenotype data, BC Safebox – a secure remote desktop environment for controlled access and collaborative research management, and BC Predict – a web service for variant interpretation, curation and reporting, designed for clinical and medical researcher uses in pathogenicity. Figure 2 below shows an example of the variant curation interface in BC Predict.
The digital cooperatives team deployed BC Platforms and the surrounding environment in a manner appropriate for sensitive genomics information. In collaboration with Monash University’s central IT (eSolutions), we contracted an external security penetration testing service to assess the deployment for handling sensitive information without losing the inherent scalability and configurability of the Monash Research Cloud. Figure 3 shows the high-level components of the ASPREE genomics information system.inherent scalability and configurability of the Monash Research Cloud. A high-level components diagram of the ASPREE genomics information system is shown in Figure 3.
After four years of operations, the genomics system continues to establish close collaborations with national and international research communities. It has produced high impact research outcomes along the way3 4 5. The R@CMon team is excited about supporting the ASPREE Genomics team as it scales up its research endeavours.
This article can also be found, published created commons here 6.
- https://aspree.org/aus/
- https://aspree.org/aus/about-the-aspree-project/sub-studies/
- Genet Med. 2020 Nov;22(11):1883-1886. doi: 10.1038/s41436-020-0881-7. Epub 2020 Jul 1. Medically actionable pathogenic variants in a population of 13,131 healthy elderly individuals. Paul Lacaze 1, Robert Sebra 2 3, Moeen Riaz 4, Jane Tiller 4, Jerico Revote 4, James Phung 4, Emily J Parker 4, Suzanne G Orchard 4, Jessica E Lockery 4, Rory Wolfe 4, Maya Strahl 2, Ying C Wang 2, Rong Chen 2 3, Daniel Sisco 3, Todd Arnold 3, Bryony A Thompson 5, Daniel D Buchanan 5 6, Finlay A Macrae 5, Paul A James 5, Walter P Abhayaratna 7, Trevor J Lockett 8, Peter Gibbs 9, Andrew M Tonkin 4, Mark R Nelson 4 10, Christopher M Reid 4 11, Robyn L Woods 4, Anne M Murray 12, Ingrid Winship 5, John J McNeil 4, Eric Schadt 2 3
- Genomic Risk Prediction for Breast Cancer in Older Women. Lacaze P, Bakshi A, Riaz M, Orchard SG, Tiller J, Neumann JT, Carr PR, Joshi AD, Cao Y, Warner ET, Manning A, Nguyen-Dumont T, Southey MC, Milne RL, Ford L, Sebra R, Schadt E, Gately L, Gibbs P, Thompson BA, Macrae FA, James P, Winship I, McLean C, Zalcberg JR, Woods RL, Chan AT, Murray AM, McNeil JJ. Cancers (Basel). 2021 Jul 14;13(14):3533. doi: 10.3390/cancers13143533.
- Rare Germline Pathogenic Variants Identified by Multigene Panel Testing and the Risk of Aggressive Prostate Cancer. Nguyen-Dumont T, Dowty JG, MacInnis RJ, Steen JA, Riaz M, Dugué PA, Renault AL, Hammet F, Mahmoodi M, Theys D, Tsimiklis H, Severi G, Bolton D, Lacaze P, Sebra R, Schadt E, McNeil J, Giles GG, Milne RL, Southey MC. Cancers (Basel). 2021 Mar 24;13(7):1495. doi: 10.3390/cancers13071495.
- Revote, Jerico; Aung, Swe Win; Quenette, Steve; Lacaze, Paul (2022): ASPREE information systems for genomic medicine using targeted panel sequencing. Monash University. Online resource. https://doi.org/10.26180/19669584.v1
