Introduction
In this series we are exploring the Nvidia BlueField 2 DPUs (Data Processing Units). We predict that before too long, DPUs will be pervasive in the datacenter, where many users will be using DPUs without realising. This series is for data centre, cloud and HPC builders, who seek to understand and master DPUs for themselves. Most will use DPUs via software some other company has written (e.g. your favourite cybersecurity software). However, some will encode some business critical function (like encryption) onto DPUs, ensuring the users have no idea (no performance loss, no need to learn anything). Check out Steve’s GTC 2021 talk – “Securing Health Records for Innovative Use with Morpheus and DPUs” for a good introduction to DPUs for this series.
For the purposes of this series, our goal is to offload encryption from virtual machines running on each host onto the DPUs. This has two important benefits:
- Eliminates the need for VM users (researchers in our context) to add transport layer security themselves, creating a lower level of entry knowledge required for them to do their work breaking down the technical barrier.
- Achieves higher work / processing throughput as the security work is offloaded from the CPU itself.
A DPU is specialised programmable hardware for data processing outside of the CPU, but still on the server. The DPUs contain their own CPU, some accelerators (e.g. for encrypting), a NIC and can be programmed to behave differently depending on your needs.
In this blog we are looking at the most basic functionality: configuring DPUs as NICs for communication between two hosts. We’ve compiled some steps and a list of some of the things that caught us out. Each of these steps were run on both hosts unless otherwise noted.
By default the DPU should act like a NIC out of the box. However it may have already been used for something else. Sometimes the DPU will be loaded with the image you want… sometimes it won’t. Hence we will assume we will need a fresh start to work from. If you are anything like us, you’re using a pair of Ubuntu 20.04.3 LTS installations running on Dell servers with a mix of brand new DPUs and older DPUs.
Glossary of terms: DOCA = Data Center-on-a-Chip Architecture DPU = Data Processing Unit NIC = Network Interface Card OVS = Open Virtual Switch (also known as Open vSwitch)
What are we trying to achieve?
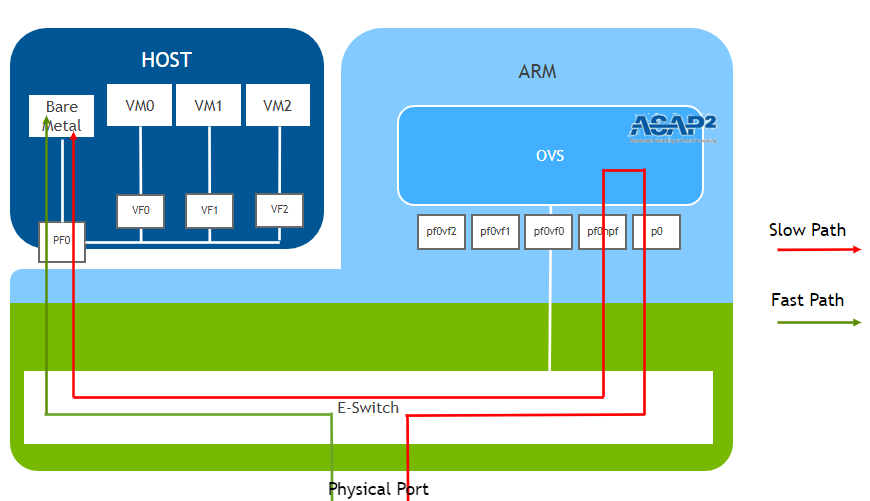
In the logical (OVS) diagram provided by Nvidia we see that inside the DPU, the physical port connects to the p0 interface which is forwarded to the pf0hp0, which then appears inside the host as the PF0 interface. In the diagram below we see two of the modes the DPU can run.
The “Fast Path” mode bypasses the DPU processors. Conversely the “Slow Path” will use the DPU’s processors. Our understanding is that all new connections first occur through the Slow Path. Then if the DPU is configured to behave as a NIC, the E-Switch knows it can bypass the DPU processors themselves. The Slow Path is the stepping stone to doing much more interesting things.
See our practical implementation for the simple DPU as a NIC case below. We keep the eth0 interfaces of the host connected to a switch for management purposes. The p1p1 (PF0) interfaces of the Bluefield 2 DPU cards are connected directly to each other.
Installing drivers, flashing the device (and installing DOCA via the NVIDIA SDK manager)
Once the DPU is installed in a PCI slot in your host machine you’ll probably want to install drivers and connect to the DPU.
DPU usually comes with Ubuntu OS installed as default. In that case we just need to install a MOFED driver on the host to be able to use the DPU.
If you want to reimage the operating system of the DPU, you will need the NVIDIA DOCA SDK installed via the NVIDIA SDK manager.
Additional information about setting up the NVIDIA SDK manager on the host can be found at: https://developer.nvidia.com/networking/doca/getting-started
In our case this meant installing the latest version of DOCA for Ubuntu 20.04.
First we downloaded the sdkmanager_1.7.2-9007_amd64.deb package and transferred it to the host. To download this file you need to be logged in to Nvidia’s dev portal so it’s best to do this from a browser)
sudo dpkg -i sdkmanager_1.7.2-9007_amd64.deb
#If you get dependency errors run the following
sudo apt-get update && sudo apt-get upgrade
sudo apt-get -f install
sudo apt-get install libxss1
sudo apt-get install docker
sudo dpkg -i sdkmanager_1.7.2-9007_amd64.deb
#Then confirm that you have the latest version with
sudo apt install sdkmanager -y
#Then run the sdkmanager
sdkmanager On the first run you will need to log in to NVIDIA’s devzone services (the sdkmanager tool prompts you to log in to a website and enter a code / scan a QR code).
We opted to use X11 forwarding to log in via the GUI.
Further information about this process can be found at:
https://docs.nvidia.com/sdk-manager/download-run-sdkm/index.html#login
Once the NVIDIA SDK manager has been installed you can install the drivers and flash the DPU using the following command:
#Note if you have a previous version of DOCA installed you can uninstall it using this command
sdkmanager --cli uninstall --logintype devzone --product DOCA --version 1.1 --targetos Linux --host
#(Re)installing DOCA
sdkmanager --cli install --logintype devzone --product DOCA --version 1.1.1 --targetos Linux --host --target BLUEFIELD2_DPU_TARGETS --flash all
#Note: Default username on the DPU is: ubuntuOn the first run you will need to log in to NVIDIA’s devzone services (the sdkmanager tool prompts you to log in to a website and enter a code / scan a QR code). Further information about this process can be found at: https://docs.nvidia.com/doca/sdk/installation-guide/index.html
Once the DPU has been successfully flashed you will need to reboot the host to ensure the new interfaces (p1p1 and p1p2) are present.
Note: We renamed the interfaces to p1p1 and p1p2 so that it is easier to remember and use in configuration management
#On both Hosts
sudo reboot
#Once they reboot check that p1p1 and p1p2 are present in
ip a You should see something like this in: ip link show
6: p1p1: <BROADCAST,MULTICAST,UP,LOWER_UP> mtu 1500 qdisc mq state UP mode DEFAULT group default qlen 1000
link/ether 0c:42:a1:e7:1e:b2 brd ff:ff:ff:ff:ff:ff
7: p1p2: <BROADCAST,MULTICAST,UP,LOWER_UP> mtu 1500 qdisc mq state UP mode DEFAULT group default qlen 1000
link/ether 0c:42:a1:e7:1e:b3 brd ff:ff:ff:ff:ff:ff
There should also be management and rshim interfaces of the DPU present.
10: enp66s0f0v0: <BROADCAST,MULTICAST> mtu 1500 qdisc noop state DOWN mode DEFAULT group default qlen 1000
link/ether ca:29:81:20:cb:51 brd ff:ff:ff:ff:ff:ff
13: tmfifo_net0: <BROADCAST,MULTICAST,UP,LOWER_UP> mtu 1500 qdisc fq_codel state UNKNOWN mode DEFAULT group default qlen 1000
link/ether 00:1a:ca:ff:ff:02 brd ff:ff:ff:ff:ff:ff
You can verify the drivers and firmware that are installed by using the command:
ethtool -i p1p1
ubuntu@HOST-17:~$ ethtool -i p1p1
driver: mlx5_core
version: 5.5-1.0.3 ← Mellanox ofed driver version
firmware-version: 24.32.1010 (MT_0000000561) ← Firmware version of DPU
expansion-rom-version:
bus-info: 0000:42:00.0
supports-statistics: yes
supports-test: yes
supports-eeprom-access: no
supports-register-dump: no
supports-priv-flags: yes
Connecting to DPU
Now that DOCA has been installed and the DPU has been flashed with the firmware, we can connect to the DPU. In our case here we configure an ip address at the rshim interface to access DPU.
ip addr add 192.168.100.1/24 dev tmfifo_net0
9: tmfifo_net0: <BROADCAST,MULTICAST,UP,LOWER_UP> mtu 1500 qdisc fq_codel state UNKNOWN group default qlen 1000
link/ether 00:1a:ca:ff:ff:02 brd ff:ff:ff:ff:ff:ff
inet 192.168.100.1/24 scope global tmfifo_net0
valid_lft forever preferred_lft forever
inet6 fe80::21a:caff:feff:ff02/64 scope link
valid_lft forever preferred_lft forever
We access DPU via ssh from the Host, but other methods of connecting are listed here: https://docs.mellanox.com/display/MFTV4120/Remote+Access+to+Mellanox+Devices
#Connect to the DPU from the Host
ssh ubuntu@192.168.100.2
#To check the driver and firmware (on the DPU)
ethtool -i p0
#Query from flint (On the DPU)
sudo flint -d /dev/mst/mt41686_pciconf0 q
#Check default OVS configuration (On the DPU)
sudo ovs-vsctl showThe query from flint should look something like this on the DPU:
ubuntu@localhost:~$ sudoflint -d /dev/mst/mt41686_pciconf0 q
Image type: FS4
FW Version: 24.32.1010
FW Release Date: 1.12.2021
Product Version: 24.32.1010
Rom Info: type=UEFI Virtio net version=21.2.10 cpu=AMD64
type=UEFI Virtio blk version=22.2.10 cpu=AMD64
type=UEFI version=14.25.17 cpu=AMD64,AARCH64
type=PXE version=3.6.502 cpu=AMD64
Description: UID GuidsNumber
Base GUID: 0c42a10300e71eb2 12
Base MAC: 0c42a1e71eb2 12
Image VSD: N/A
Device VSD: N/A
PSID: MT_0000000561
Security Attributes: N/AThe default config of OVS should look something like this on the DPU:
ubuntu@localhost:~$ sudo ovs-vsctl show
10c2d713-1ca3-4106-8eea-1178f3c1348d
Bridge ovsbr1
Port p0
Interface p0
Port pf0hpf
Interface pf0hpf
Port ovsbr1
Interface ovsbr1
type: internal
Bridge ovsbr2
Port p1
Interface p1
Port pf1hpf
Interface pf1hpf
Port ovsbr2
Interface ovsbr2
type: internal
ovs_version: "2.14.1"
Thanks to the default OVS (Open Virtual Switch) configuration you can also add IP addresses to the p1p1 interfaces on the hosts to enable connection between them.
#On Host-16
sudo ip addr add 10.10.10.16/24 p1p1
sudo ip link set p1p1 up#On Host-17
sudo ip addr add 10.10.10.17/24 p1p1
sudo ip link set p1p1 up
ping -I p1p1 10.10.10.16Troubleshooting:
If your OVS configuration does not match the example or the ping test fails, you might want to try removing all existing OVS configurations using the “ovs-vsctl del-br“
For example if you had a bridge called “arm-ovs” you could delete it with the following command
sudo ovs-vsctl del-br arm-ovsThen recreate the default OVS bridges ovsbr1 and ovsbr2 with the following commands:
ovs-vsctl add-br ovsbr1
ovs-vsctl add-port ovsbr1 pf0hpf
ovs-vsctl add-port ovsbr1 p0
# configure p1p2 (optional in our case, p1p2 is not used)
ovs-vsctl add-br ovsbr2
ovs-vsctl add-port ovsbr2 pf1hpf
ovs-vsctl add-port ovsbr2 p1
ip link set dev ovsbr1 up
ip link set dev ovsbr2 upImportant note: Adding p0 and p1 of DPU to the same ovs bridge could cause a loop and potentially create a multicast issue in the network.
Some observations, noting we’re dealing with very new technology:
- Installing DOCA sdk via the command line is not yet simple.
- Sometimes DOCA install may fail and trying it again usually works. (Note: The newest version “1.2.0” does not seem to have this issue)
- To be able to use both ports of the DPU, we observe they need to be configured with IP addresses of the different vlan on the host.
Conclusion
Once the DPUs have been installed and flashed correctly you can easily add IP addresses to the p1p1 interfaces on the hosts to enable network configuration. In the next post we’ll look at the NVIDIA DOCA East-West Overlay Encryption Reference Application.
Written by Ben Boreham, Swe Aung and Steve Quenette as part of a partnership between Nvidia, the Australian Research Data Commons (ARDC), and Monash University
