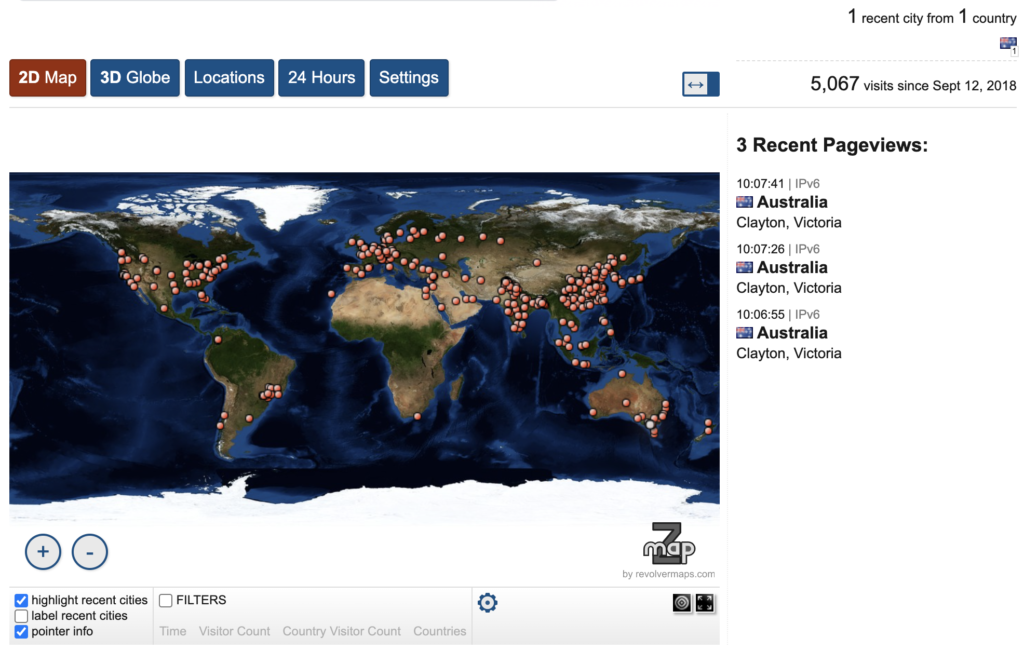
Dr. Kelly Wyres is a research fellow in the Holt Lab. Kelly first approached the Research Cloud at Monash team in 2019. She sought assistance to migrate their bioinformatics web application – Kaptive to Monash infrastructure. Kaptive is a user-friendly tool for finding known loci within one or more pre-assembled genomes, specifically for the identification of Klebsiella surface polysaccharide loci. It presents these results in a novel and intuitive web interface, helping the user to rapidly gain confidence in locus matches. Kaptive has been developed and currently maintained by Kelly Wyres, Ryan Wick and Kathryn Holt at Monash University. It also uses bacterial reference databases that are carefully curated by Kelly Wyres and Johanna Kenyon from Queensland University of Technology.
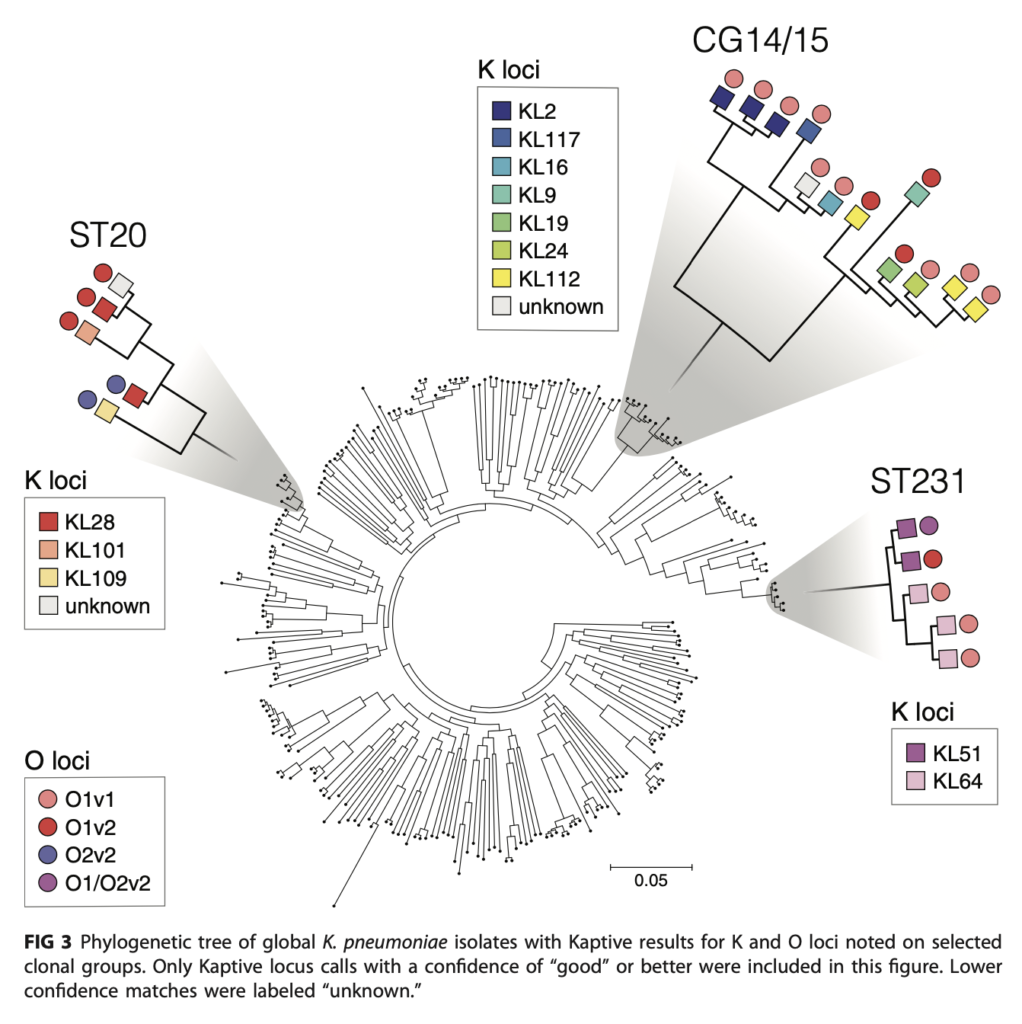
The R@CMon team provided its standard LAMP platform to host Kaptive on the Research Cloud. This included helping Kelly transition Kaptive from its original web2py mechanism (to quickly create web applications), to a production grade LAMP stack including a dedicated web server and storage backend. Now transitioned, the team can efficiently and effectively cooperate with Kaptive alongside a critical mass of other domain-specific LAMP based applications across all disciplines of research. The team also assisted in applying additional security controls (e.g HTTPS/SSL, reCAPTCHA) on the server to improve its security posture. As a measure of impact, more than 3000 searches (and associated computing jobs) have been submitted into Kaptive to date. As new reference databases become ready and curated, it’ll then be incorporated into Kaptive and made available to the research community.
This article can also be found, published created commons here 1.