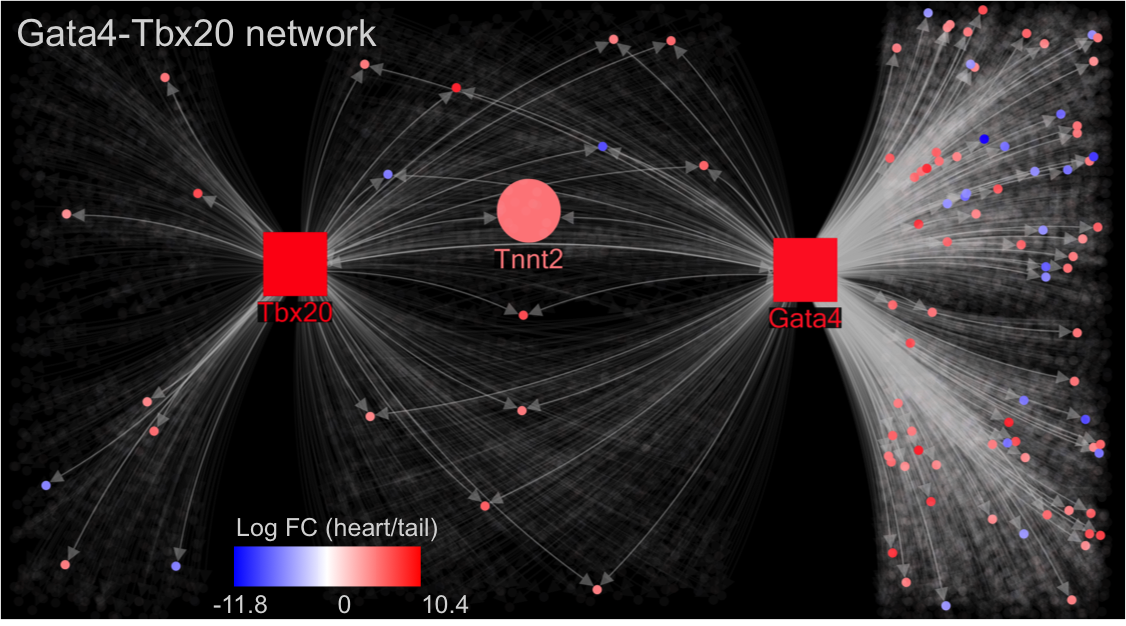
Back in early 2014, the R@CMon team assisted SBI Australia to deploy the VISIONET (Visualizing Transcriptomic Profiles Integrated with Overlapping Transcription Factor Networks) visualisation web service on the Monash node of the NeCTAR Research Cloud. Since then, VISIONET has been further enhanced to support more complex transcription factor network topologies. To date, VISIONET has been published in two papers.
The R@CMon team will continue supporting SBI Australia with its plan to further develop the VISIONET web service this year.