Interferons (IFNs) were identified as antiviral proteins more than 50 years ago. However, their involvement in immunomodulation, cell proliferation, inflammation and other homeostatic process has since been identified. These cytokines are used as therapeutics in many diseases such as chronic viral infections, cancer and multiple sclerosis. These IFNs regulate the transcription of approximately 2000 genes in a IFN subtype, dose, cell type and stimulus dependent manner.

Interferome Wordle
Interferome is an online database of IFN regulated genes. The database is a valuable resource for biomedical researchers, being regularly used by scientists from across the world. This database of IFN regulated genes is an attempt at integrating information from high-throughput experiments to gain a detailed understanding of IFN biology. Interferome enables reliable identification of an individual Interferon Regulated Gene (IRG) or IRG signatures from high-throughput data sets (i.e. microarray, proteomic data etc.). It also assists in identifying regulatory elements, chromosomal location and tissue expression of IRGs in humans and mice.
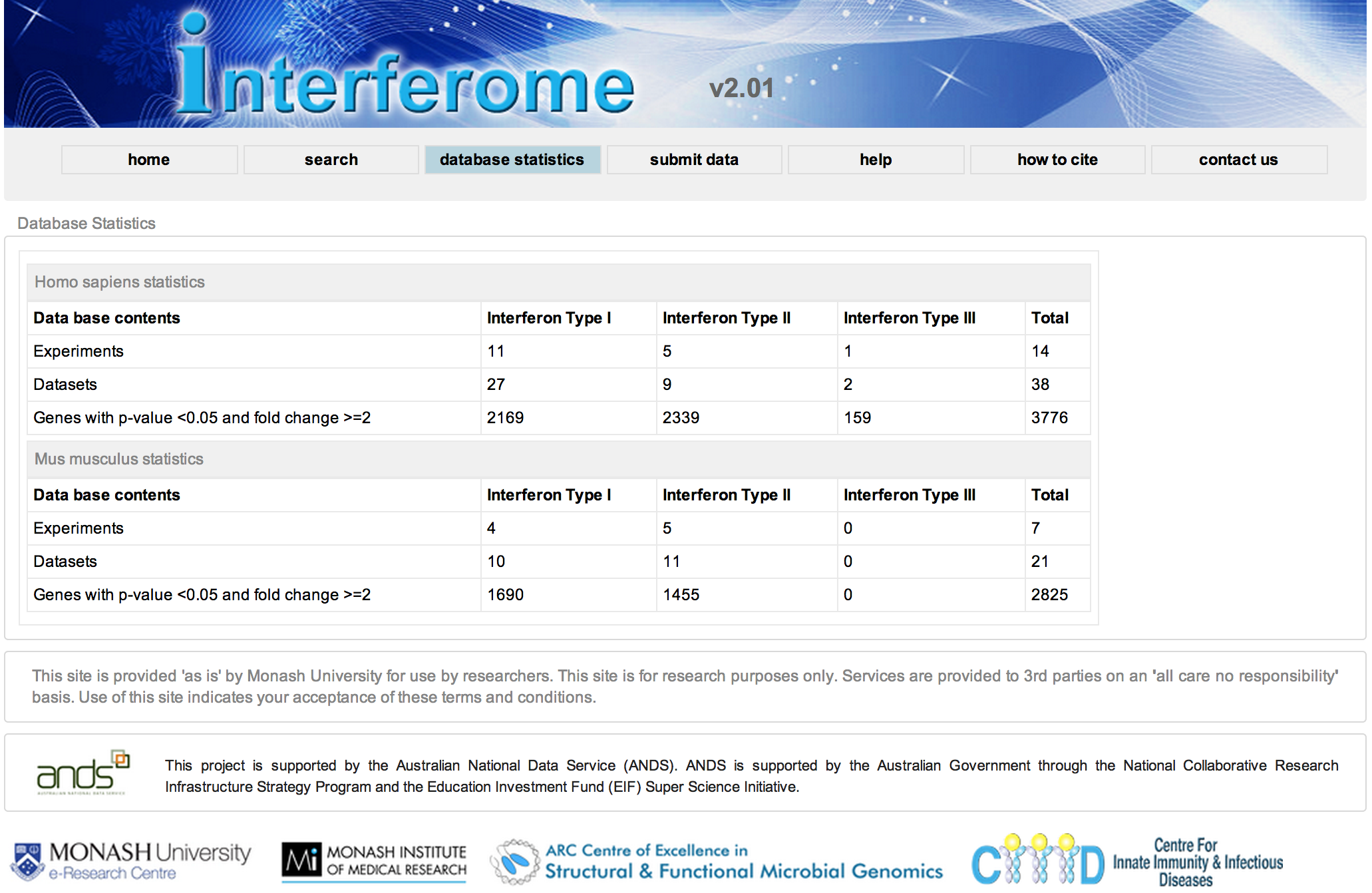
Interferome Database Statistics
The R@CMon team assisted Prof. Paul Hertzog and the Centre of Innate Immunity & Infectious Diseases at MIMR-PHI in migrating versions 1.0 and 2.0 of the Interferome online database into the NeCTAR Research Cloud. Interferome Version 2.0 has quantitative data, more detailed annotation and search capabilities and can be queried for one gene or thousands as in a gene list from a microarray experiment. To ensure availability of data and assist researchers with hypothesis generation and novel biological discoveries, the Interferome database is backed by VicNode Collection 2014R9.06. More information about Interferome is available on the help page.