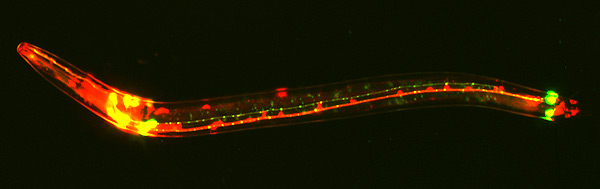
Measuring the changes in gene expressions levels and determining differential expressed genes during the processes of human immunodeficiency virus (HIV) infection, replication and latency is instrumental in further understanding HIV infections. These measurements or studies are vital in developing strategies for virus eradication from the human body. Dr. Chen Li, a research fellow from the Immunoproteomics Laboratory at Monash University has developed a novel compendium of comprehensive functional genes annotations from genes expressions and proteomics studies. The genes in the compendium have been carefully curated and shown to be differentially expressed during HIV infection, replication and latency.

The HIVed Online Database, Front Page
The R@CMon team assisted with the deployment of the online database – HIVed on the Monash node of the NeCTAR Research Cloud. The system has been running on R@CMon and serving the public community for more than a year. HIVed is considered to be the first fully comprehensive database that combines datasets from a wide range of experimental studies that have been carefully curated using a variety of experimental conditions. The datasets are further enriched by integrating it with other public databases to provide additional annotations for each data points. The HIVed online database has been developed to facilitate the functional annotation and experimental hypothesis HIV related genes with an intuitive web interface which enables dynamic display or presentation of common threads across HIV latency and infection conditions and measurements. The work done for the development of HIVed has been recently published into Scientific Reports and the Immunoproteomics Laboratory has plans to incorporate new experimental studies and external annotations into the HIVed database as they become available.