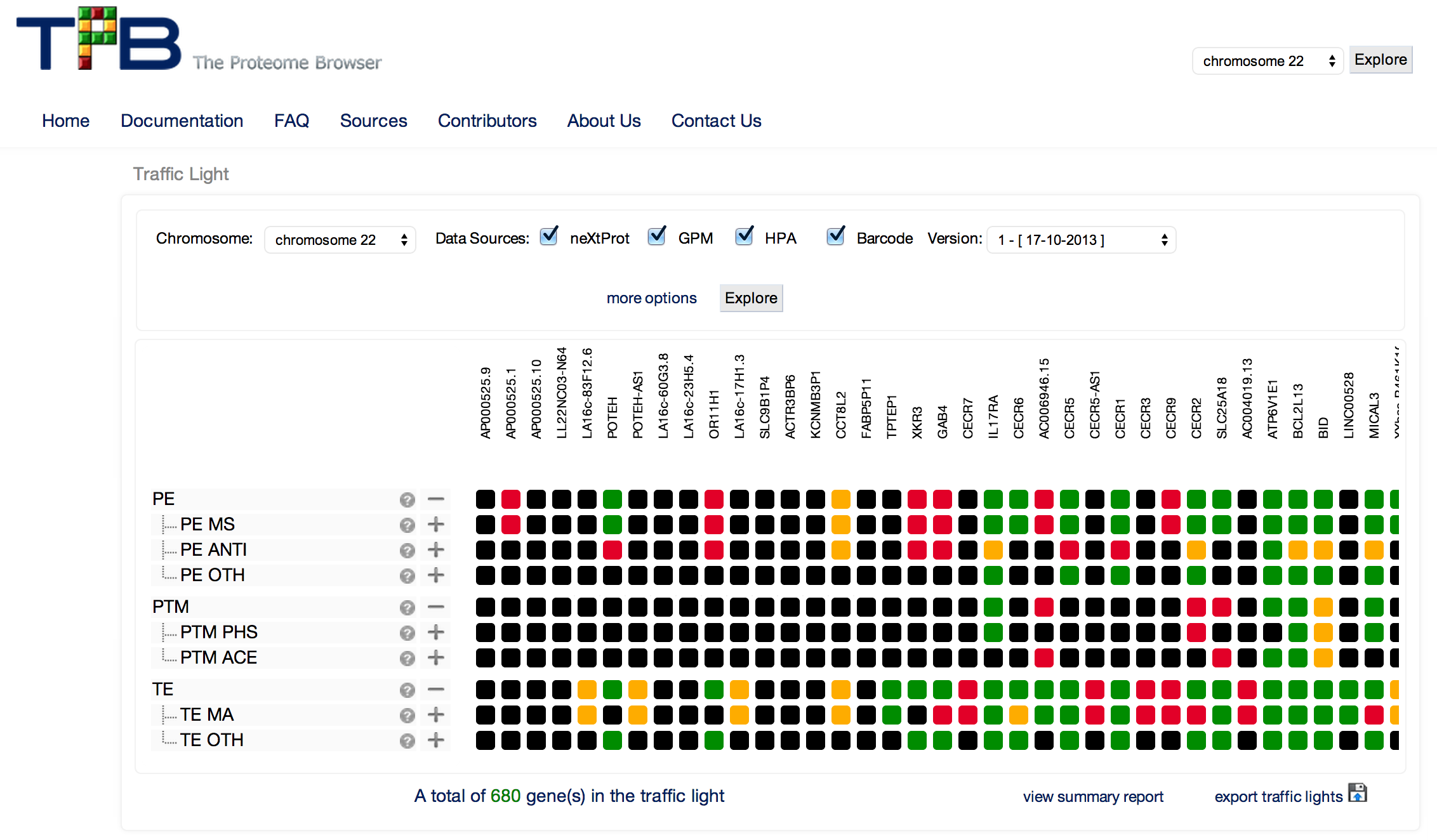
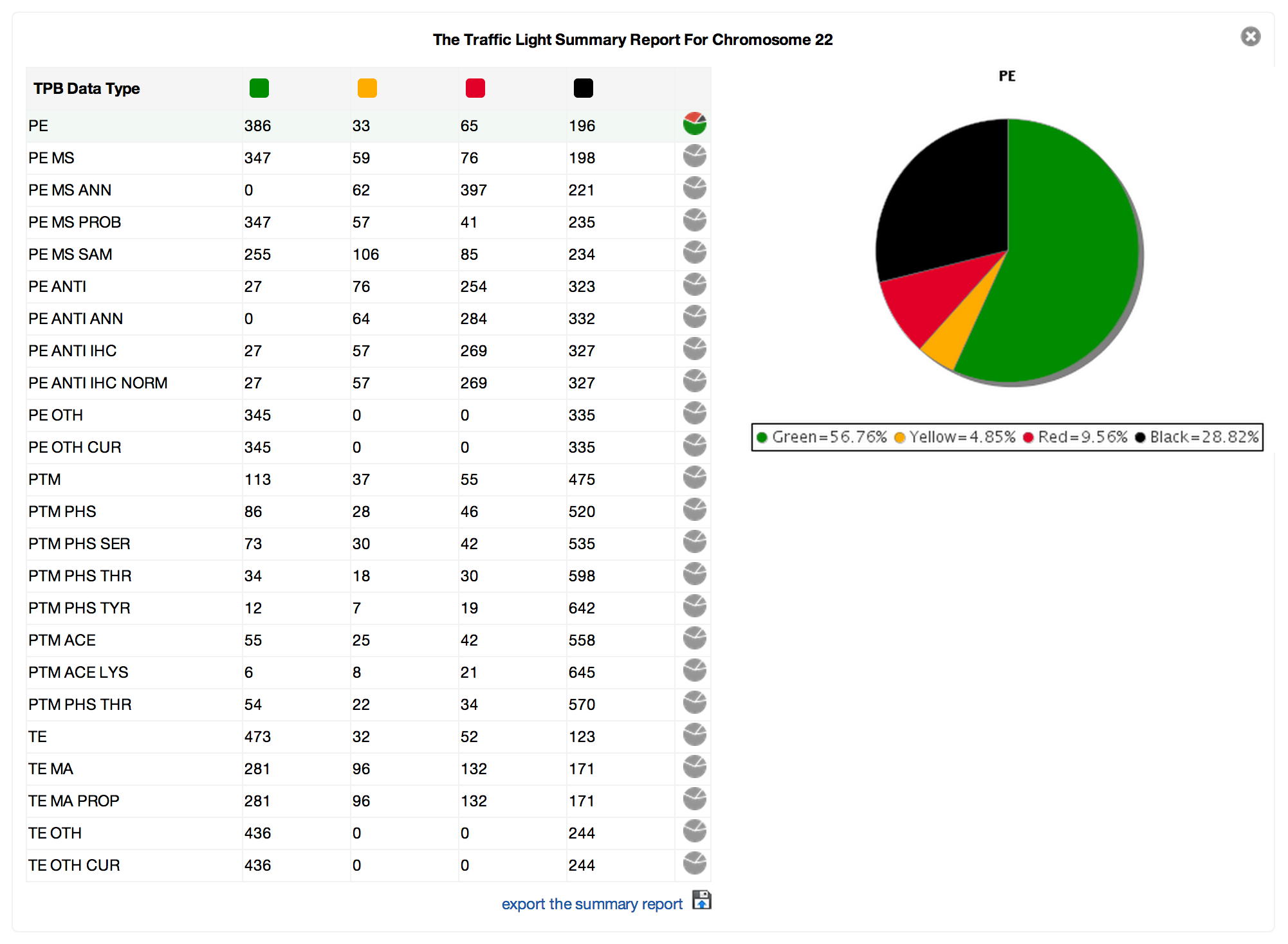
The Proteome Browser (TPB) is a web portal that integrates human protein data and information. It provides an up-to-date view of the proteome (the entire library of proteins that can be expressed by cells or organisms – like us!) across large gene sets to support human proteome characterisation as part of the Chromosome-centric Human Proteome Project (C-HPP). Pertinent genomic and protein data from multiple international biological databases are assembled by TPB in a searchable format supporting C-HPP’s global proteomics effort.
TPB’s framework extracts biological data from numerous sources, maps it into the genome, and performs categorisation on the results based on quality and information content. The result (level of evidence) is presented by TPB using a simple point matrix coded by traffic light system (green – highly reliable evidence, yellow – reasonable evidence, red – some evidence is available or black – there is no available evidence). TPB uses hierarchical data types to group similar information from different experiment types.
TPB is supported by Monash University, Monash eResearch Centre (MeRC), Chromosome-centric Human Proteome Project (C-HPP), Australia/New Zealand Chromosome 7 Consortium and the Australian National Data Service (ANDS). Researchers are now using TPB in various proteomic-related discoveries.
The R@CMon cloud team recently provided assistance to migrate The Proteome Browser web service to be hosted on the Monash node of the NeCTAR Research Cloud. TPB is using persistent storage (Volumes) granted via a VicNode computational storage allocation to house its underlying database. TPB’s new home will ensure it has stable and scalable long-term hosting supported by the NeCTAR and RDSI federal research infrastructure programmes.