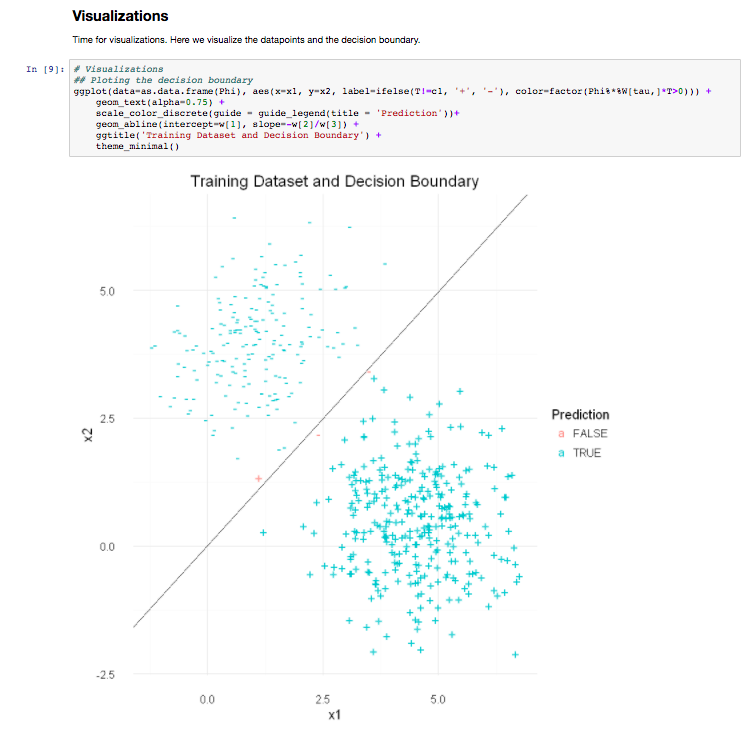
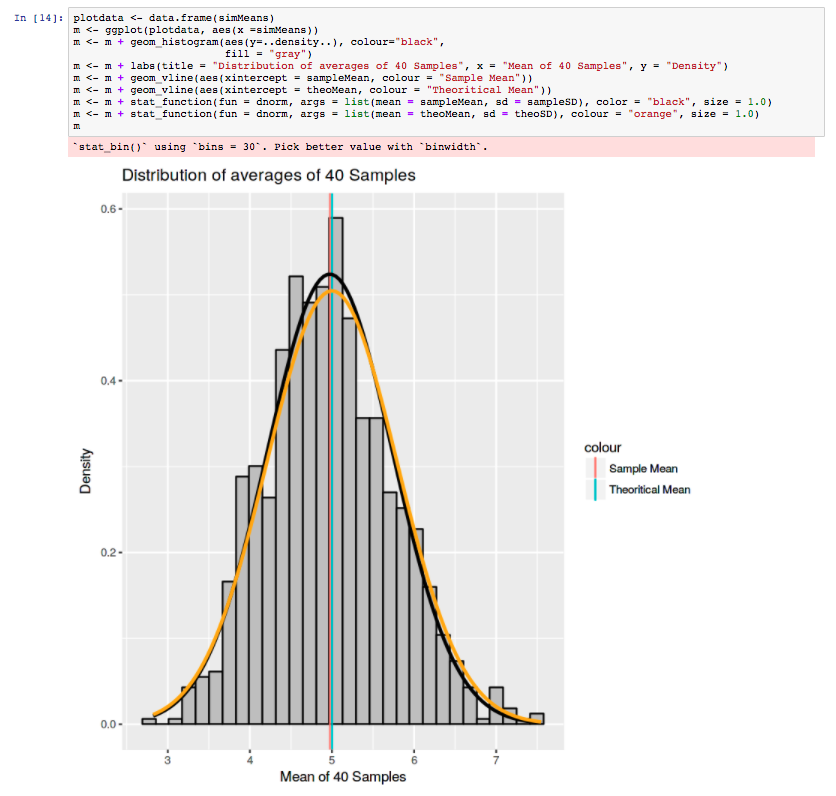
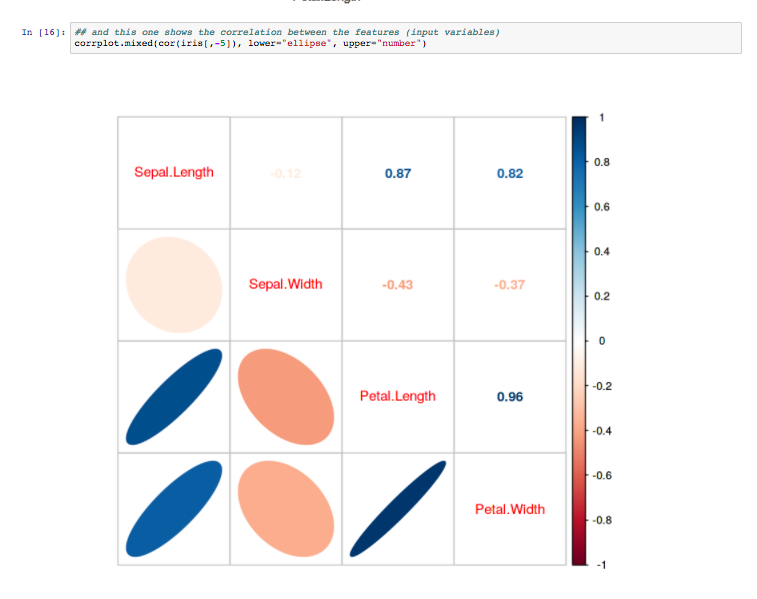
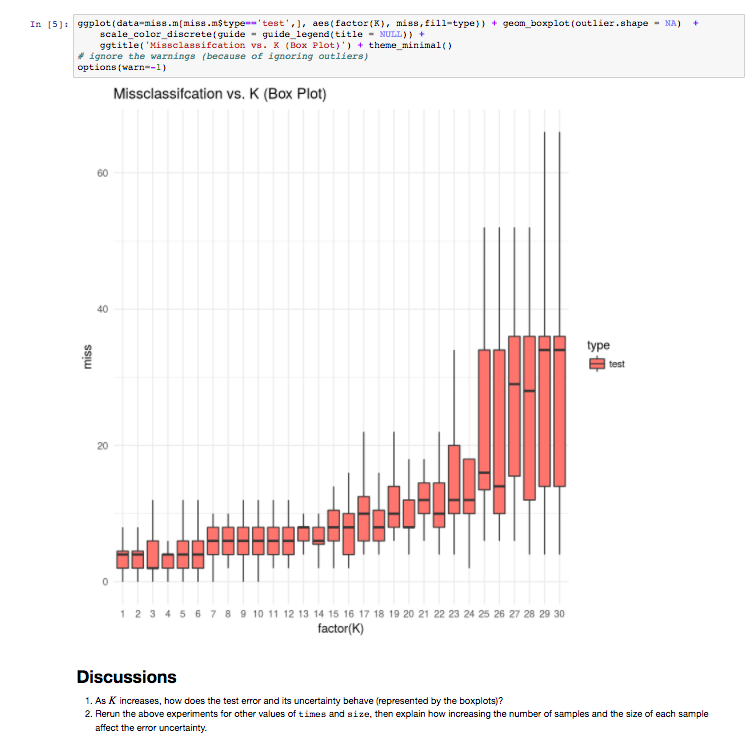
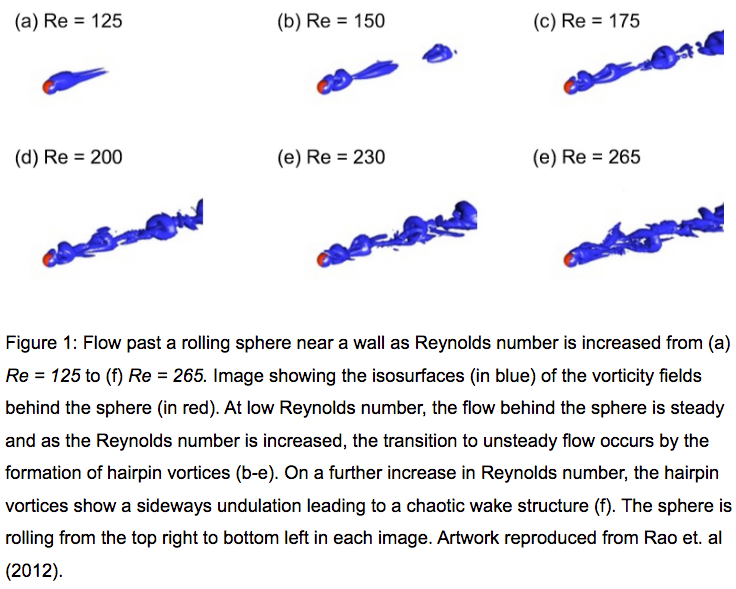
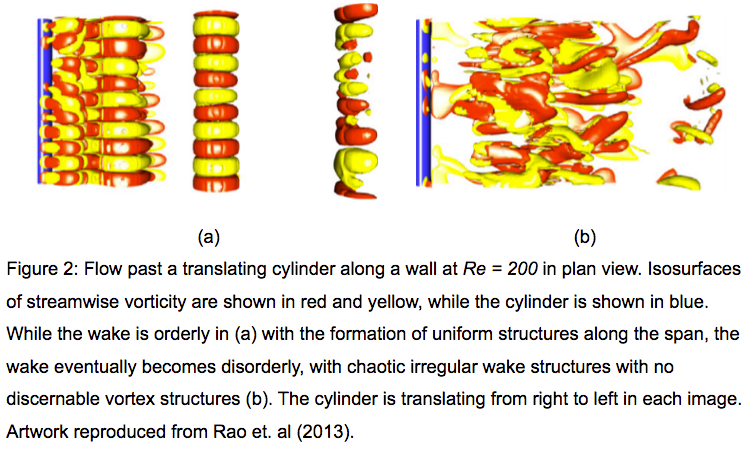
The Monash Country Lines Archive (MCLA) is a collaborative project between the Monash Indigenous Centre (MIC), Faculty of Arts and the Faculty of Information Technology with a team of researchers, digital animators and students. The MCLA aims to support the indigenous Australian communities in the preservation of stories that combine their history, knowledge, poetry, songs, performance and language. MCLA began working with the Yanyuwa people of Borroloola, NT, creating a number of animations between 2007 and 2010. It was these animations that caught the attention of Dr Alan Finkel, the then Chancellor of Monash University. In 2011, the Alan and Elizabeth Finkel Foundation supported the project for a further five years.

Render from “Why We All Die” 2015 ©MCLA & Taungurung Dolodanin-dat Animation Group.
Since its foundation in 2011, the MCLA has produced nine short-form animated films ranging from four minutes to twenty-four minutes in length while working with the communities through every step of the animation process; script, storyboards, character and landscape concepts and construction, animation, rendering, sound and post-production. Initially, producing these animations was challenging due to their heavy computational requirements. The MCLA team didn’t have access to any dedicated render-farm resources as would be normal for a commercial animation studio, so all rendering works were done on individual desktops and laptops. This resource limitation forced the MCLA team to compromise advanced rendering techniques in order to quickly render a large number of scenes while still maintaining a certain level of production quality.

Render frame from “Jibi the Giant Spirit Birds” 2013 ©MCLA & Nyamba Buru Yawuru.
In 2013, the MCLA team gained access to the NeCTAR Research Cloud, giving them a much needed rendering capacity boost. The R@CMon team assisted the MCLA in deploying and dynamically scaling their workflow into a distributed rendering workflow in the research cloud. Modelling, animation and rendering software have been licensed and configured on this virtual render farm. The farm has been configured so that MCLA can easily access it remotely to submit jobs and inspect their renders. The MCLA then started applying advanced rendering techniques in their workflow, techniques that weren’t possible on their previous setup. After several years of usage, demands for MCLA to produce more and more high quality visualisations also increased. This required the render farm to scale more, much more, and it did.

Render frame from “Janyju the Red Lizard” 2014 ©MCLA & Nyamba Buru Yawuru.
Access to the research cloud-backed render farm removed a huge limitation for the MCLA, inspiring them to produce more animations for the indigenous Australian communities without compromising on quality. The R@CMon team will continue to support the MCLA going forward and will be there when the time comes that the farm needs more power. The MCLA is composed of Dr John Bradley, Dr Shannon Faulkhead, Brent D McKee, Dr Tom Chandler and Chandara Ung.
-

-
Render from “How Man Found Fire” 2016 ©MCLA & Taungurung Dolodanin-dat Animation Group.
-

-
Render from “How Man Found Fire” 2016 ©MCLA & Taungurung Dolodanin-dat Animation Group.
-

-
Render frames from “Emu Hunters of Excellence” 2015 ©MCLA & Garrwa Families.
-

-
Render frame from “The Groper” 2013 ©MCLA & Yanyuwa Families.
-

-
Render frames from “Emu Hunters of Excellence” 2015 ©MCLA & Garrwa Families.
-

-
Render from “How Man Found Fire” 2016 ©MCLA & Taungurung Dolodanin-dat Animation Group.
-

-
Render from “How Man Found Fire” 2016 ©MCLA & Taungurung Dolodanin-dat Animation Group.
-

-
Render frames from “Why We All Die” 2015 ©MCLA & Taungurung Dolodanin-dat Animation Group.
-

-
Render frame from “The Groper” 2013 ©MCLA & Yanyuwa Families.
-

-
Render frames from “Emu Hunters of Excellence” 2015 ©MCLA & Garrwa Families.
-

-
Render frame from “The Groper” 2013 ©MCLA & Yanyuwa Families.
-

-
Render frames from “Emu Hunters of Excellence” 2015 ©MCLA & Garrwa Families.
-

-
Render frame from “The Groper” 2013 ©MCLA & Yanyuwa Families.
-

-
Render frame from “The Groper” 2013 ©MCLA & Yanyuwa Families.
-

-
Render frames from “Why We All Die” 2015 ©MCLA & Taungurung Dolodanin-dat Animation Group.
-

-
Render frames from “Why We All Die” 2015 ©MCLA & Taungurung Dolodanin-dat Animation Group.
-

-
Render frame from “The Groper” 2013 ©MCLA & Yanyuwa Families.