FLAIR (Fluids Laboratory for Aeronautical and Industrial Research), from the Department of Mechanical and Aerospace Engineering, Faculty of Engineering, has been conducting experimental and computational fluid mechanics research for over twenty years, focusing on fundamental fluid flow problems that impact the automotive, aeronautical, industrial and biomedical fields.
A key research focus in recent years has been understanding the wake dynamics of particles near walls. Particle-particle and wall-particle interactions were investigated using an in-house spectral-element numerical solver. Understanding these interactions is key in many engineering industries. When applied to biological engineering, blood cells / leukocytes are numerically modelled as canonical bluff bodies (i.e., as cylinders and spheres) and numerical computations are carried out. These simulations are not only useful in understanding biological cell transport but have wider applications in mineral processing, chemical engineering and applications in ball sports. Due to the computational and data-intensive nature of this research, it has always been a challenge to get access to sufficient computing resources for its needs.
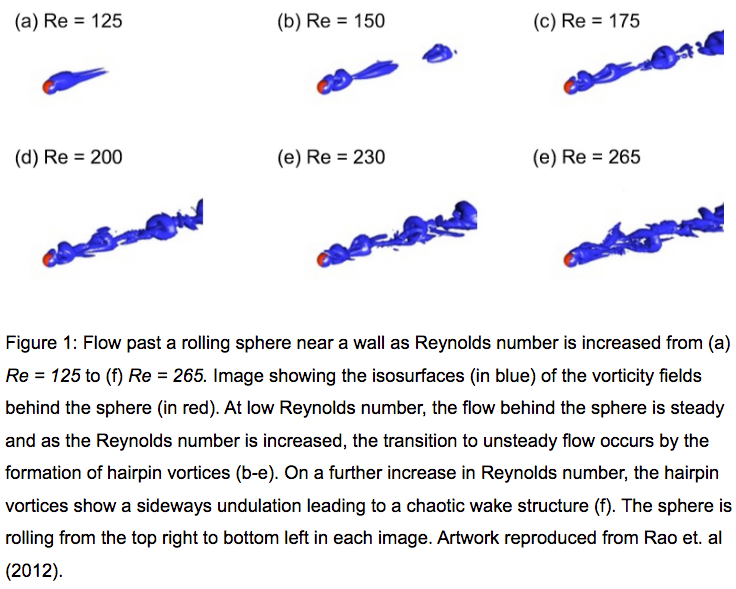
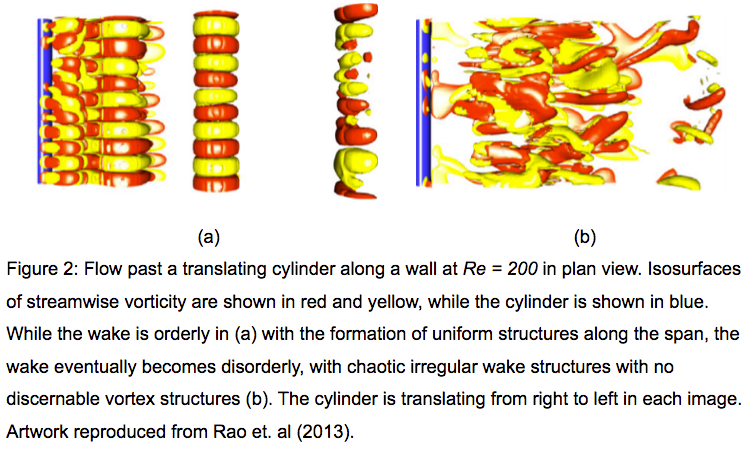
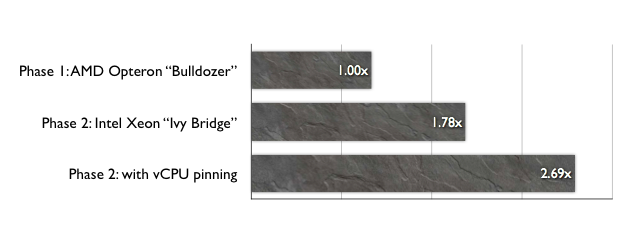
In particular, their project aims to understand the wake dynamics on multiple particles in various scenarios such as rolling, collisions and vortex-induced vibrations; and the resultant mixing which occurs as a result of these interactions, etc. The group’s two- and three-dimensional fluid flow solver also incorporates two-way body dynamics to model these effects. As the studies involve multiple parameters such as Reynolds number, body rotation, height of the body above the wall, etc, the total parameter space is extensive, requiring significant computational resources. While the two-dimensional simulations are carried out on single processors, their three-dimensional counterparts require parallel processing, making NeCTAR nodes an ideal platform to run these computations. Some of the visualisations from the group’s three-dimensional simulations are shown in Figures 1 and 2 below.
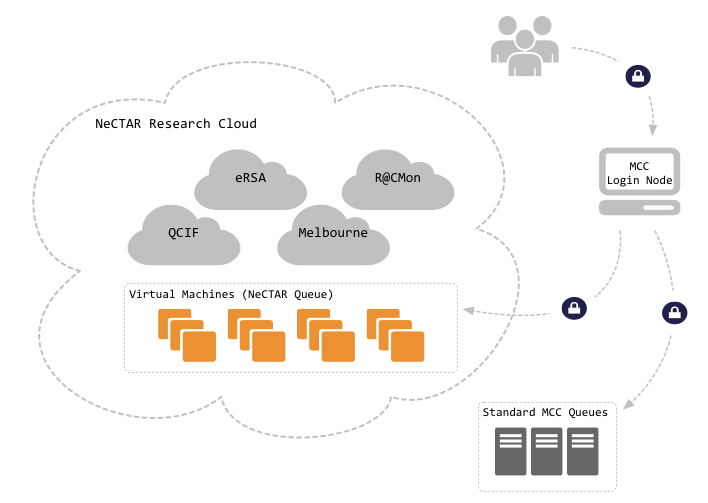
Since 2008, the FLAIR team has been making good use of the Monash Campus Cluster (MCC), a high-performance/high-throughput heterogeneous system with over two thousand CPU cores. However, MCC is heavily utilised by researchers from across the university and FLAIR users often found themselves waiting long periods before they could run their fluid flow simulations. It became clear that FLAIR researchers needed additional computational resources.
R@CMON was able to secure a 160-core allocation to the FLAIR team, which increased valuable resources for the group. Now, thanks to both NeCTAR and MCC-R@CMon, over one million CPU hours distributed across 4,000 jobs were provided for the project’s CPU-intensive calculations.
This has resulted in a number of publications in the highest impact fluid mechanics journals, with several more in a pre-submission stage; for example:
- Rao, A., Thompson, M.C., & Hourigan, K. (2016) “A universal three-dimensional instability of the wakes of two-dimensional bluff bodies.” Journal of Fluid Mechanics, 792, 50-66.
- Rao, A., Radi, A., Leontini, J.S., Thompson, M.C., Sheridan, J., & Hourigan, K. (2015) “A review of rotating cylinder wake transitions.” Journal of Fluids and Structures, 53, 2–14.
- Rao, A., Radi, A., Leontini, J.S., Thompson, M.C., Sheridan, J., & Hourigan, K. (2015) “The influence of a small upstream wire on transition in a rotating cylinder wake.” Journal of Fluid Mechanics (published online) 769 (R2), 1-12. DOI
- Rao, A., Thompson, M.C., Leweke, T., & Hourigan, K. (2013) “The flow past a circular cylinder translating at different heights above a wall.” Journal of Fluids and Structures, 41, 9–21.
- Rao, A., Passaggia, P.-Y., Bolnot, H., Thompson, M.C., Leweke, T., & Hourigan, K. (2012) “Transition to chaos in the wake of a rolling sphere.” Journal of Fluid Mechanics, 695, 135-148.