The Australian Bureau of Statistics (ABS) provides public access to internet activity data as “data cubes” under the catalog number “8153.0”. These statistics are derived from data provided by internet service providers (ISPs) and offer an estimate of the number of users (frequency) having access to a specific Internet technology such as ADSL. While this survey is adequate for general observations, the granularity is too coarse to assess the impact of internet access on Australian society and economic growth. The Geodata Server project led by Klaus Ackermann (Faculty of Business and Economics, Monash University) was created with an aim to provide significantly enhanced granularity on Internet usage, in both the temporal and spatial dimensions for Australia on a local government level and for other cities worldwide.
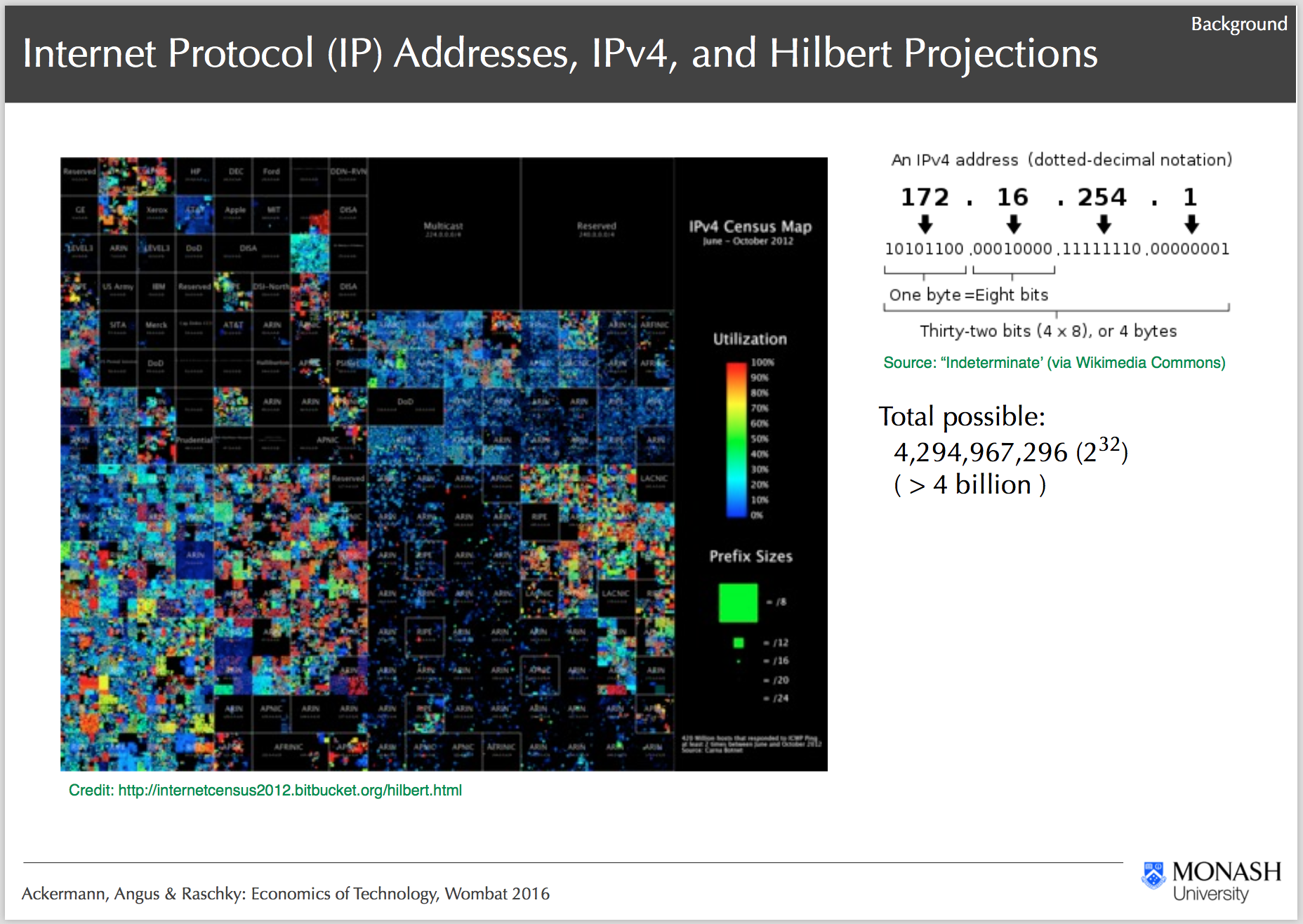
IPv4 Heatmap and Project Background, Ackermann, Angus & Raschky: Economics of Technology, Wombat 2016
One of the main challenges in the project is the analysis of 1.5 trillion observations from the ABS data sets. The project requires high-performance and high-throughput computational resources to analyse this vast amount of data. Also, a reasonable amount of data storage space is vital for storing reference and computed data. Another major challenge is how to architect the analysis pipeline to fully utilise the available resources. Over the last 3 years, several iterations of the methodology as well as infrastructure setup have been developed and tested to optimise the analysis pipeline. The R@CMon team engaged with Klaus to address the various computational, storage and analysis requirements of the project. A dedicated NeCTAR project has been provisioned for Geodata Server, which includes the computational resources to be used on the Monash node of the NeCTAR Research Cloud. Computational storage was provisioned to the project via VicNode allocation scheme.
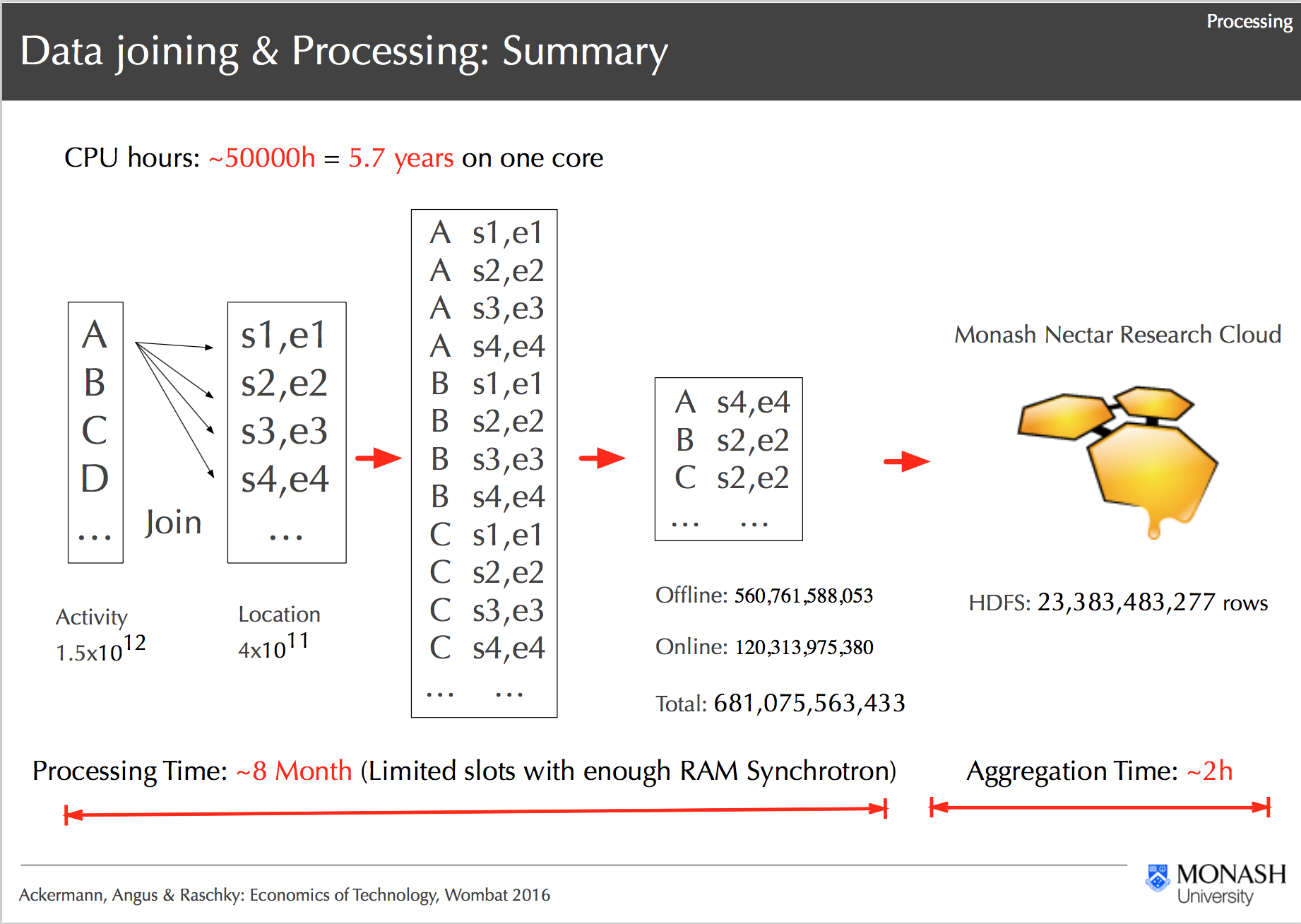
Processing Workflow on R@CMon, Ackermann, Angus & Raschky: Economics of Technology, Wombat 2016
With the computational and storage resources in place, the project was able to progress with the development of the analysis pipeline based on various “big data” technologies. In coordination with the R@CMon team, several Hadoop distributions have been evaluated, namely, Cloudera, MapR and Hortonworks. The latter was chosen for its ease of installation and 100% open source commitment. The resulting cluster consists of 32 cores with 8TB of Hadoop Filesystem (HDFS) storage divided among 4 nodes. Tested configuration includes 16 cores and 2 nodes or 32 cores and 1 node. The data has been distributed into 2TB volume drives. The master node of the cluster has an extra large volume attached to store the raw (reference) data. To optimize the performance of the distributed HDFS, all loaded data is stored in compressed Lempel–Ziv–Oberhumer (LZO) format to reduce the burden on the network, that is shared among other tenants on the NeCTAR Research Cloud.
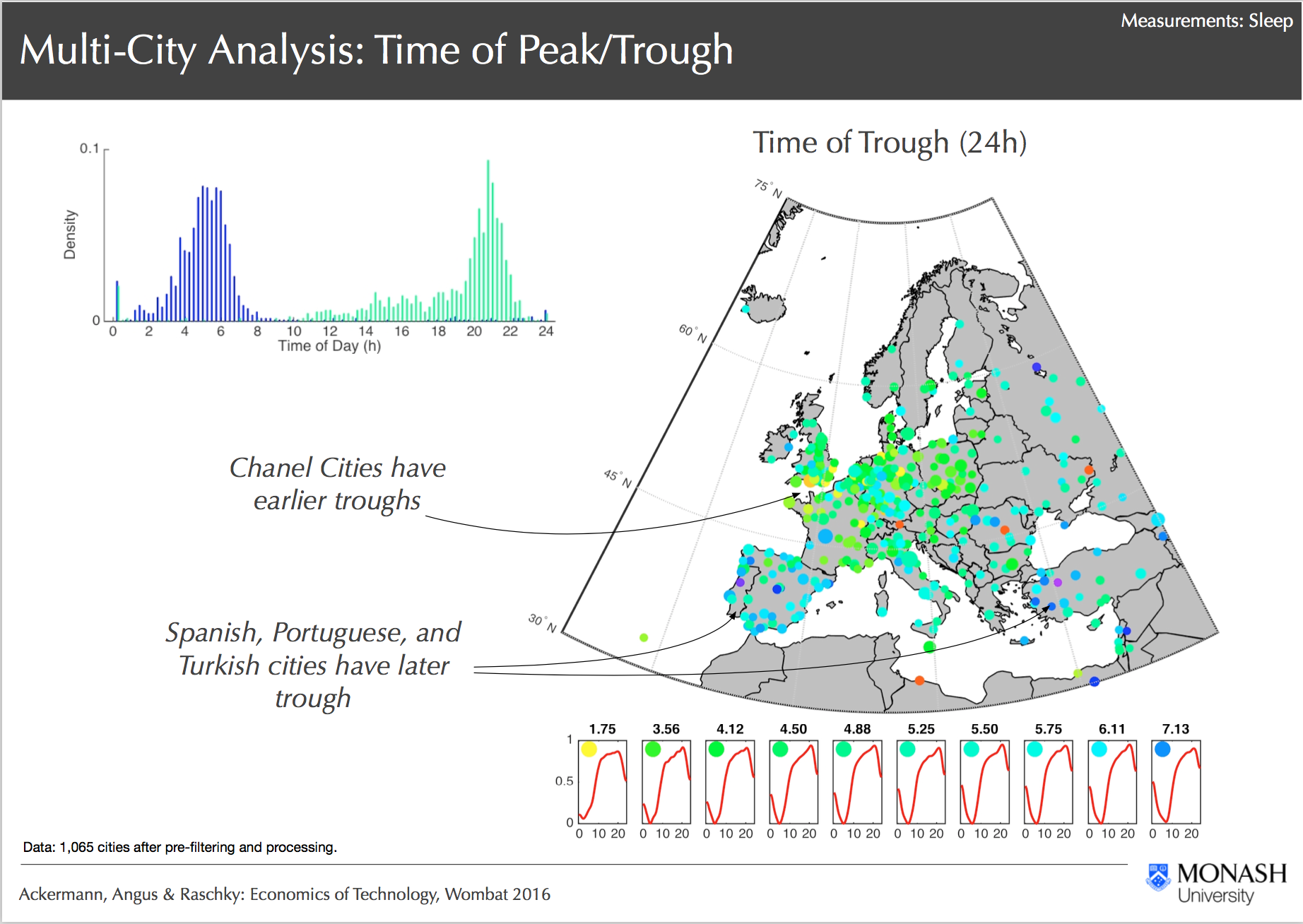
Multi City Analysis, Ackermann, Angus & Raschky: Economics of Technology, Wombat 2016
Through R@CMon, the Geodata Server project was able to successfully handle and curate trillions of IP-activity observational data and link these data accurately to its geo-location in single and multi-city models. Analysis tools were laid down as part of the pipeline from high-performance (HPC) processing on Monash’s supercomputers, to Hadoop-like type of data parallelisation in the research cloud. From this, preliminary observations suggest strong spatial-correlation and evidence of political boundaries discontinuities on IP activities, which suggests some cultural and/or institutional factors. Some of the models produced from this project are currently being curated in preparation for public release to the wider Australian research community. The models are actively being improved with additional IP statistical data from other cities in the world. As the data grows, the analysis pipeline, computational and storage requirements are expected to scale as well. The R@CMon team will continue to support the Geodata Server project to reach its next milestones.