We’re pleased to announce we now have GPGPU accelerated cloud instances working, a first for the NeCTAR Research Cloud!
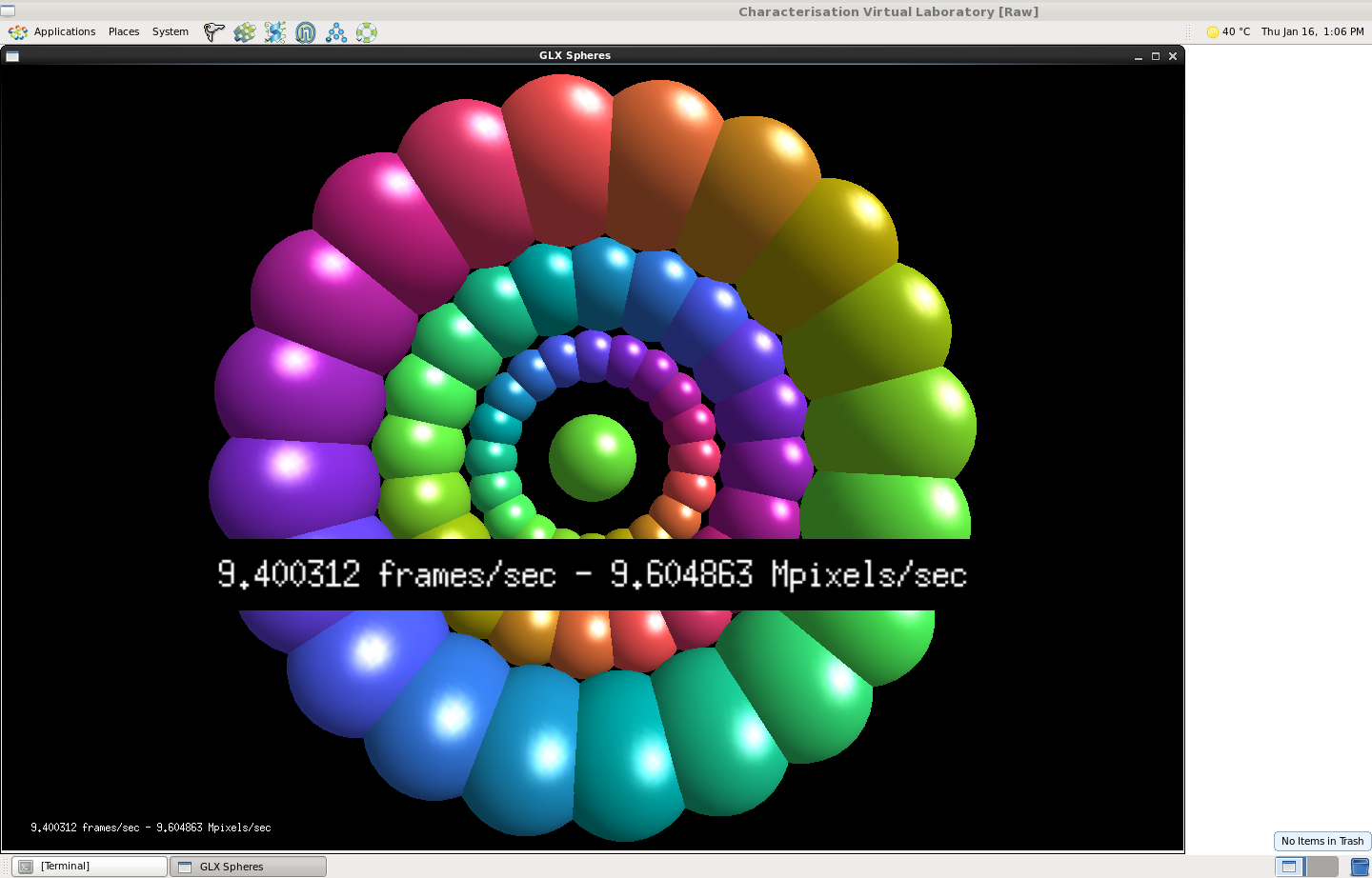
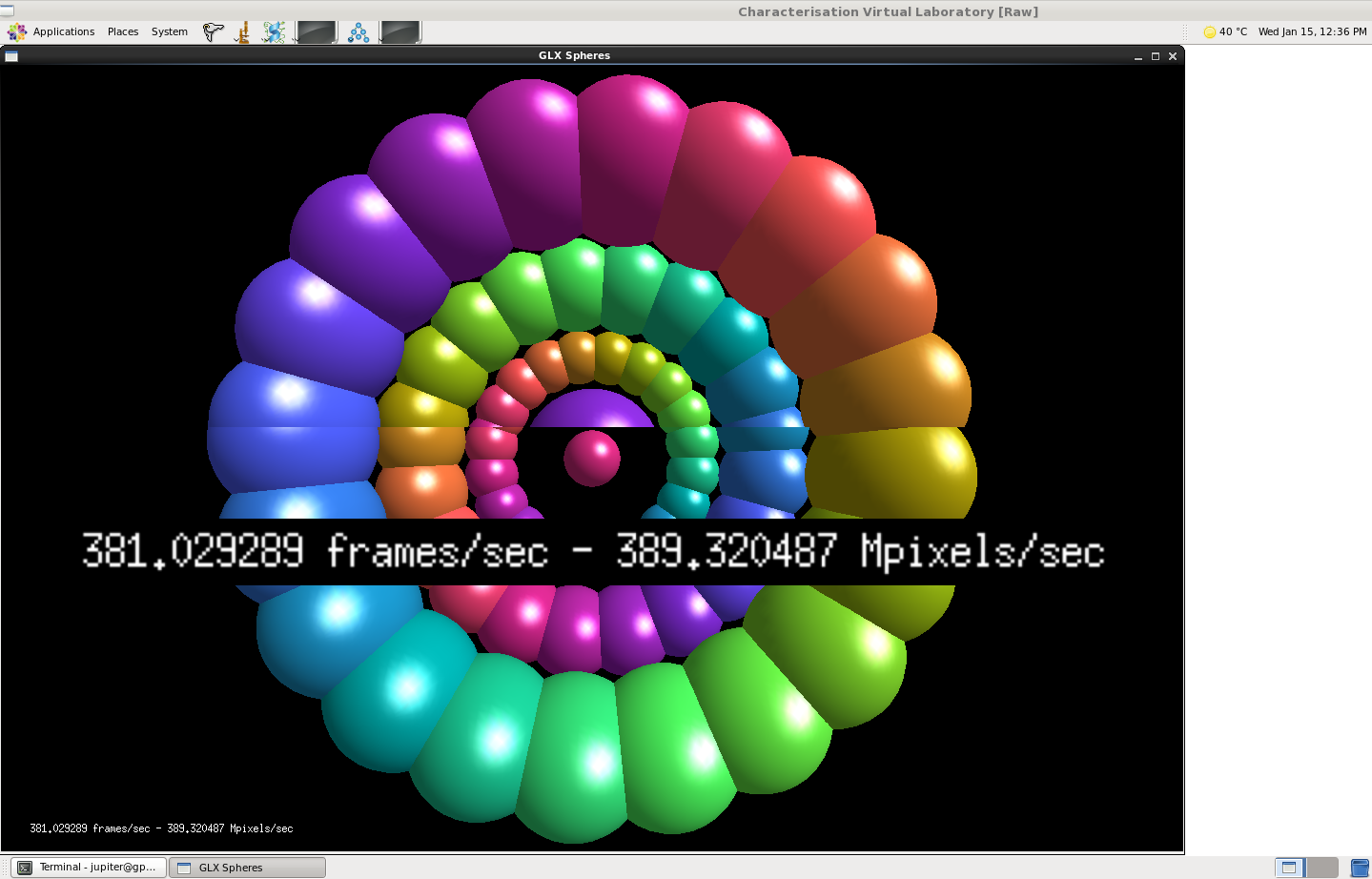
If you’re not as excited by SSH session logs as I am you might like the following screenshots captured by Juptier Hu from the CVL team, which illustrate the CVL running on both a new GPU flavor and a standard non-GPU flavor. The CVL uses TurboVNC to achieve remote hardware accelerated rendering (care of VirtualGL), so the framerates here show CPU versus GPU rendering speeds.
And to really prove it, here’s the obligatory mono-spaced console dump you’d expect from a technical blog:
01:47:51 blair@bethwaite:~$ nova show f44cad73-40e1-4e83-8699-dd3f7f2e9ead | grep flavor | flavor | cg1.medium (19) | 01:47:54 blair@bethwaite:~$ nova show f44cad73-40e1-4e83-8699-dd3f7f2e9ead | grep network | monash-test network | 11x.xxx.255.20 | 01:48:39 blair@bethwaite:~$ sshrc root@11x.xxx.255.20 root@ubuntu:~# lshw | grep -C 2 NVIDIA description: 3D controller product: GF100GL [Tesla M2070-Q] vendor: NVIDIA Corporation physical id: 6 bus info: pci@0000:00:06.0 root@ubuntu:~# nvidia-smi Mon Jan 13 01:52:22 2014 +------------------------------------------------------+ | NVIDIA-SMI 5.319.76 Driver Version: 319.76 | |-------------------------------+----------------------+----------------------+ | GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC | | Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. | |===============================+======================+======================| | 0 Tesla M2070-Q Off | 0000:00:06.0 Off | 0 | | N/A N/A P0 N/A / N/A | 10MB / 5375MB | 0% Default | +-------------------------------+----------------------+----------------------+ +-----------------------------------------------------------------------------+ | Compute processes: GPU Memory | | GPU PID Process name Usage | |=============================================================================| | No running compute processes found | +-----------------------------------------------------------------------------+ root@ubuntu:~# cd NVIDIA_CUDA-5.5_Samples/7_CUDALibraries/MersenneTwisterGP11213/ root@ubuntu:~/NVIDIA_CUDA-5.5_Samples/7_CUDALibraries/MersenneTwisterGP11213# ./MersenneTwisterGP11213 ./MersenneTwisterGP11213 Starting... GPU Device 0: "Tesla M2070-Q" with compute capability 2.0 Allocating data for 2400000 samples... Seeding with 777 ... Generating random numbers on GPU... Reading back the results... Generating random numbers on CPU... Comparing CPU/GPU random numbers... Max absolute error: 0.000000E+00 L1 norm: 0.000000E+00 MersenneTwister, Throughput = 3.1591 GNumbers/s, Time = 0.00076 s, Size = 2400000 Numbers Shutting down...
Currently these are available as cg1.* flavors in the monash-test cell, so not open for general consumption – and they won’t be for a while until we purchase and deploy more GPU nodes. The allocation process also needs to be tweaked to deal with this new capability. So currently GPU flavors are only accessible by special arrangement with the R@CMon team.
We’ll be deploying a considerable GPU capability as part of R@CMon in order to support, e.g., GPU accelerated rendering and GPGPU accelerated viz processing for the CVL. GPU capabilities can also be useful for on-demand development work, such as providing hardware rendering for the CAVE2 development environment.
At the moment we have a handful of NVIDIA Tesla M2070-Q’s, with Kepler K2’s coming as part of R@CMon phase2. If you’re keen to get access or try this out then drop us a line.