The Characterisation Virtual Laboratory (CVL) is a powerful platform that integrates Australian imaging facilities with computational and data storage infrastructure, together with sophisticated processing and analysis toolsets. The CVL platform provides scientists working in various fields with a common analysis and collaboration environment, the CVL turns the humble remote desktop into a highly flexible Scientific Software as-a-Service delivery platform powered by the NeCTAR Research Cloud.
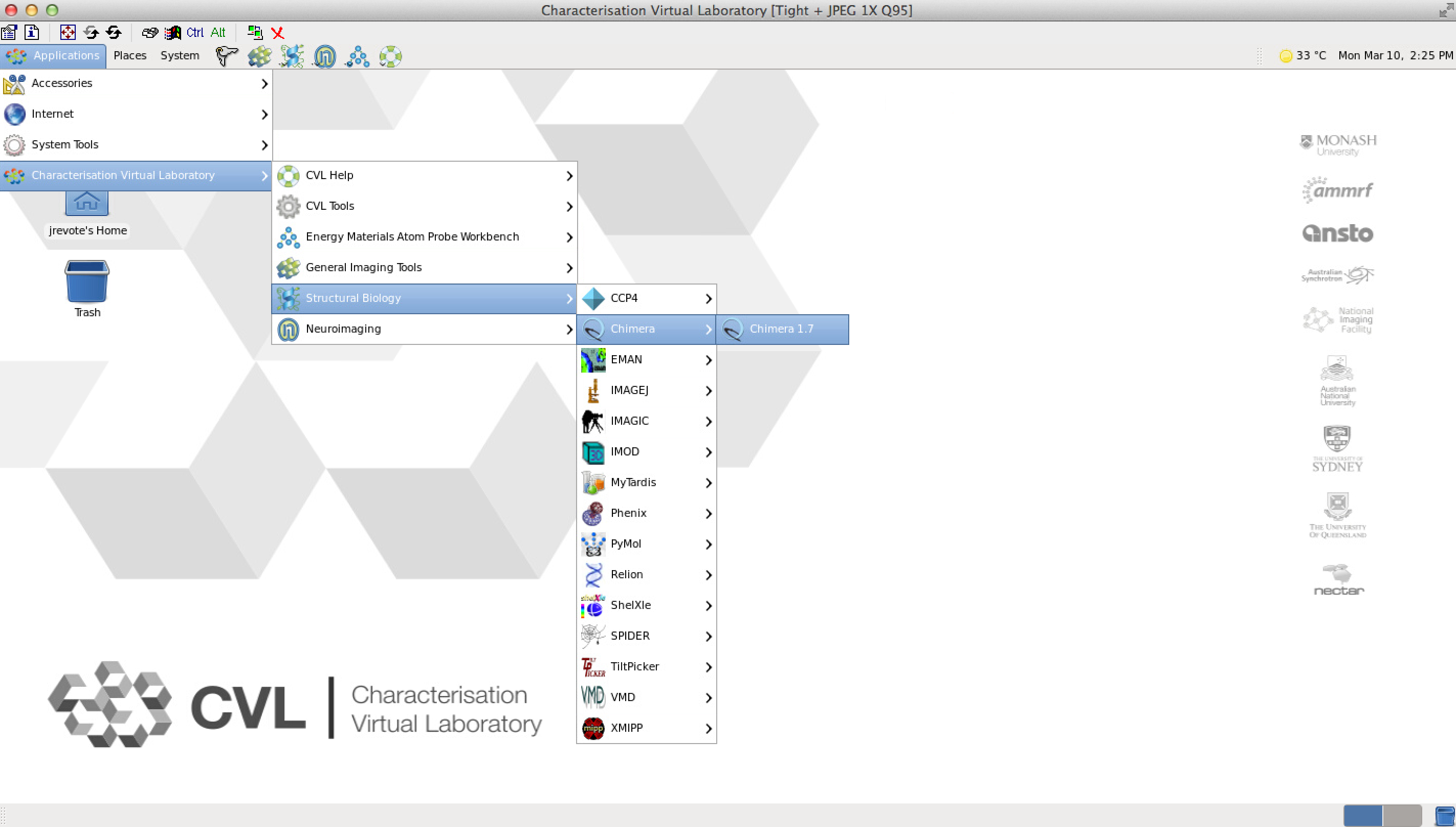
The current production CVL includes toolsets covering Neuroimaging, Energy Materials and Structural Biology research drivers. The project includes so-called “CVL fabric services”, which provide the necessary infrastructure to modularise popular software toolsets from any number of domains.
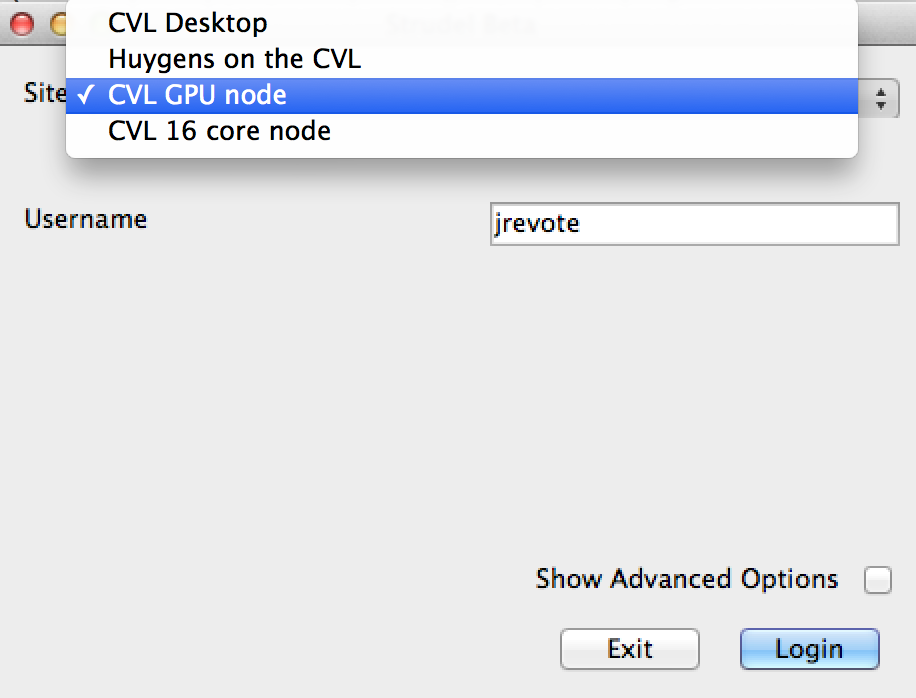
The R@CMon team assisted the CVL team in migrating CVL services into R@CMon. The use of persistent storage (Volumes on R@CMon) ensured consistent user home directories and software-stack repositories. The default “CVL Desktop” pool is now serving users with software-rendered CVL environments running on R@CMon. The CVL team is also a beta user of GPU flavours on R@CMon and is currently testing GPU-enabled CVL environments on the “CVL GPU node” pool (via CVL Launcher).
The following video demonstrates a GPU-enabled CVL environment launched on R@CMon. It shows the PyMOL and UCSF Chimera applications from the Structural Biology workbench, running and utilising the available GPU. The use of GPU enables seamless interaction and manipulation of datasets.
The plan is to increase the “CVL GPU node” pool to accommodate more users once GPU node capacity on R@CMon has been upgraded with deployment of R@CMon Phase 2. Watch this space for more CVL on R@CMon news. Other updates about the CVL and its sub-projects are also available on the CVL site.