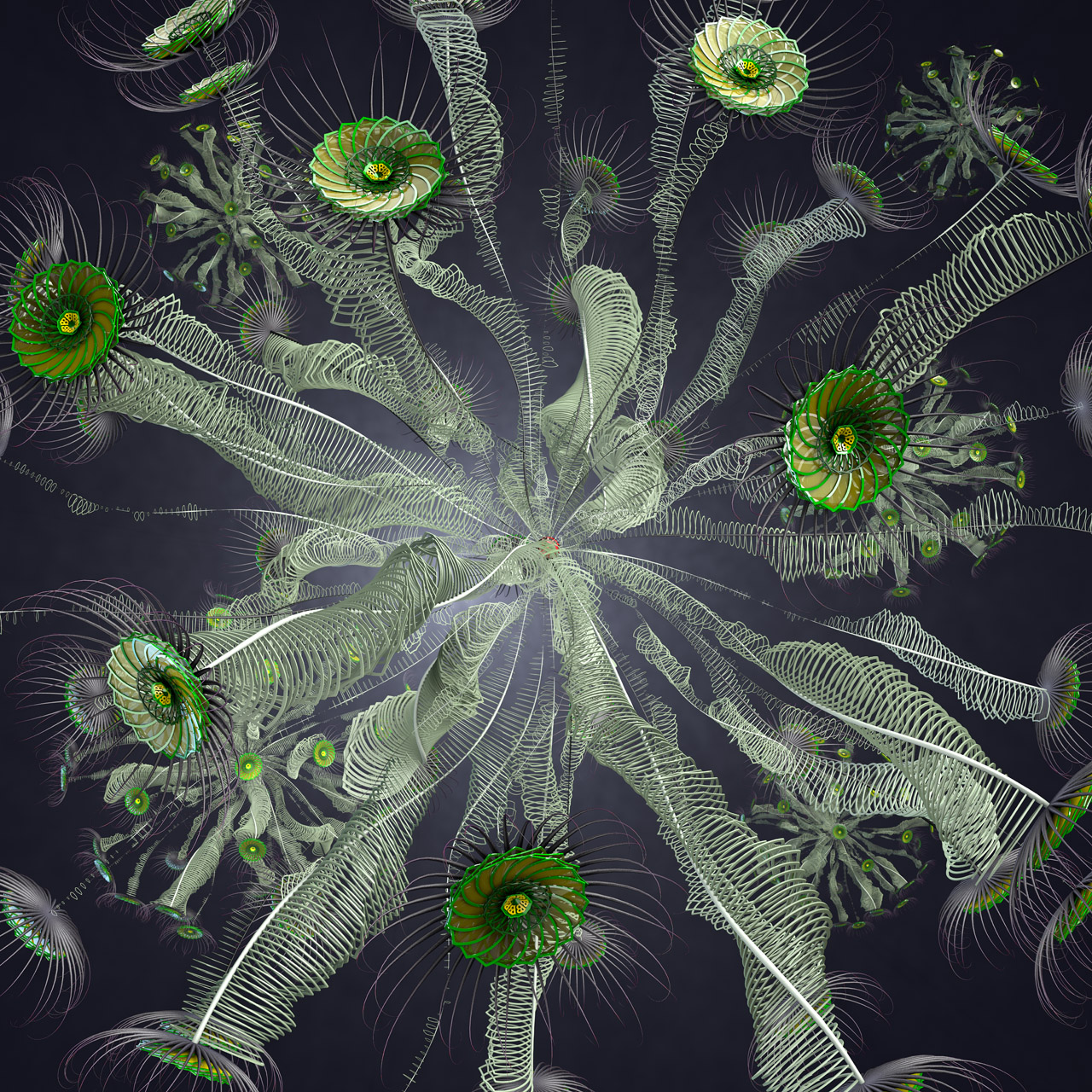
Jon McCormack, an Associate Professor from Faculty of Information Technology at Monash University, creates high-resolution renderings of various subjects. His artworks have been showcased in leading galleries, museums and symposia.
Before the advent of the Monash node of the NeCTAR Research Cloud, Jon has been limited to running his render jobs to a couple of machines he can get his hands on in the faculty. After the first phase of the Monash node went online, the R@CMon team helped Jon in porting his rendering workflow in the NeCTAR Research Cloud environment. Jon uses Cinema4D for creating his high-resolution renders. Cinema4D runs on Windows and OS X so we’ve prepared a suitable Windows Server 2012 image for this purpose.
We assessed the performance of a NeCTAR guest running a rendering job. For this, we’ve used CINEBENCH from MAXON, a well known benchmarking tool to measure and compare CPU and graphics performance of various systems and platforms. The following video shows CINEBENCH running on a 16-core guest in the Monash node.
The CINEBENCH result showed that the guest performed well on the “Main Processor Performance (CPU)” component. Once GPUs become available in the second phase of the Monash node, we’ll be looking at running the second component of CINEBENCH which measures “Graphics Card Performance (GPU)”.
Cinema4D comes with a distributed rendering system which allows unlimited render clients connecting to a render server. The render server is where render jobs are submitted and is in charge of distributing it to the available render clients. NeCTAR guests have been provisioned to be render clients in Jon’s tenancy that talks to Jon’s render server running on a dedicate machine. Each render client renders a frame or a tile and submits the finished render back to the render server.
The following are some low-res renders from the “Image from Fifty Sisters, commission for the Ars Electronica Museum, Linz, Austria 2012/2013. Copyright Jon McCormack” project that Jon produced using his render farm in the NeCTAR Research Cloud.