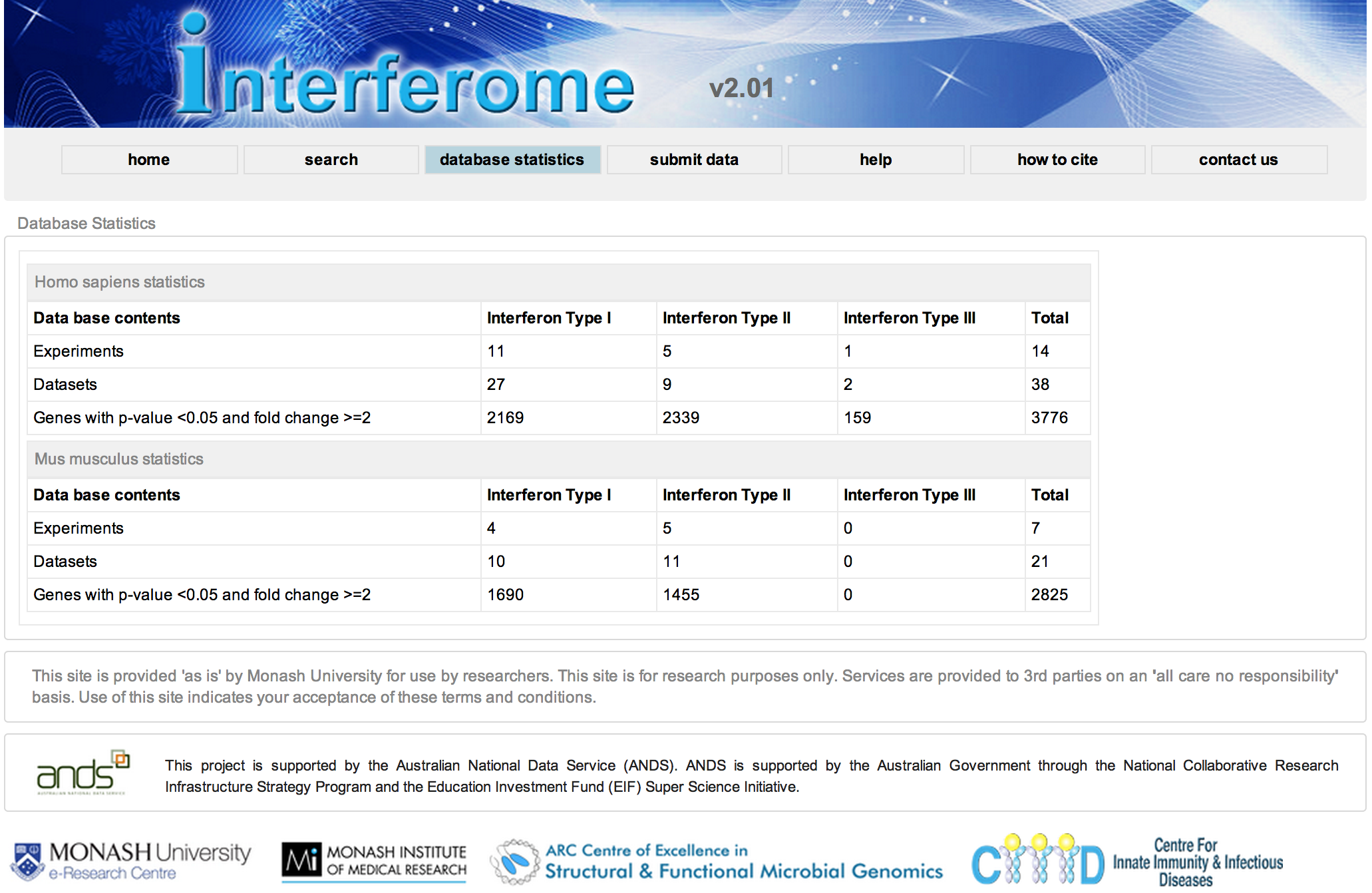
R@CMon, thanks to the Monash eResearch Centre’s long history of establishing “the right hardware for research”, prides itself on effectiveness at computing, orchestrating and storing for research. In this post we highlight an engagement that didn’t yield an “effectiveness” to our liking, and how that helped shape elements of the imminent R@CMon phase2.
In the latter part of 2013 the R@CMon team was approached by a visiting student working at the Water Sensitive Cities CRC. His research project involved parameter estimation for an ill-posed problem in ground-water dynamics. He had setup (perhaps partially inherited) an Excel spreadsheet based Monte Carlo engine for this, with a front-end sheet providing input and output to a built in VBA macro for the grunt work – an erm… interesting approach! This had been working acceptably in the small, as he could get an evaluation done within 24 hours on his desktop machine (quad core i7). But now he needed to scale up and run 11 different models, and probably a few times each to tweak the inputs. Been there yourself? This is a very common pattern!
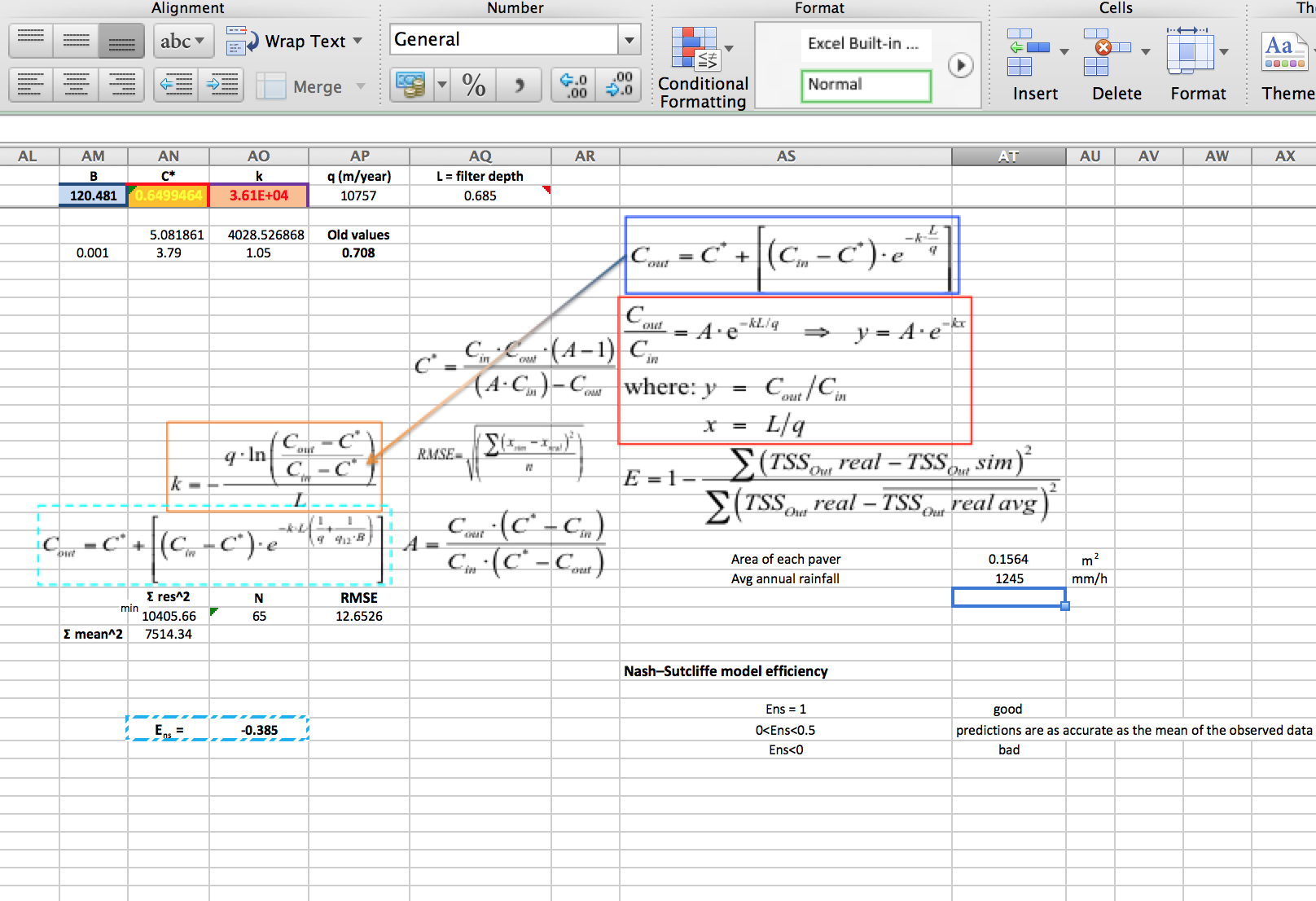
Nash-Sutcliffe model efficiency (Figure courtesy of eng. Antonello Mancuso, PhD, University of Calabria, Italy)
MCC (the Monash Campus Cluster), our first destination for ‘compute’, doesn’t have any Windows capability, and even if it did, attempting to run Excel in batch mode would have been something new for us. No problem we thought, we’ll use the RC, give him a few big Windows instances and he can spread the calculations across them. Not an elegant or automated solution for sure, but this was a one-off with some tight time constraints, so it was more important to start calculations than get bogged down with a nicer solution.
It took a few attempts to get Windows working properly. We eventually found the handy cloudbase solutions trial image and its guidance documentation. But we also ran into issues activating Windows against the Monash KMS, turns out we had to explicitly select our local network time source as opposed to the default time.windows.com. We also found some problems with the CPU topology that Nova was giving our guests, Windows was seeing multiple sockets rather than multiple cores, which meant desktop variants were out as they would ignore most of the cores.
Soon enough we had a Server 2012 instance ready for testing. The user RDP’d in and set the cogs turning. Based on the first few Monte Carlo iterations (out of the million he needed for each scenario) he estimated it would take about two days to complete a scenario, quite a lot slower than his desktop but still acceptable given the overall scale-out speed up. However, on the third day after about 60 hours compute time he reported it was only 55% complete. Unfortunately that was an unsustainable pace – he needed results within a fortnight – and so with his supervisor they resolved to code and use a different statistical approach (using PEST) that would be more amenable to cluster-computing.
We did some rudimentary performance investigation during the engagement and didn’t find any obvious bottlenecks, the guest and host were always very CPU busy, so it seemed largely attributable to the lesser floating point capabilities of our AMD Bulldozer CPUs. We didn’t investigate deeply in this case and no doubt there could be other elements at play here (maybe Windows was much slower for compute on KVM than Linux), but this is now a pattern we’ve seen with floating point heavy workloads across operating systems and on bare metal. Perhaps code optimisations for the shared FPU in the Bulldozer architecture can improve things, but that’s hardly a realistic option for a spreadsheet.
The AMDs are great (especially thanks to their price) for general purpose cloud usage, that’s why the RC makeup is dominated by them and why commercial clouds like Azure use them. But for R@CMon’s phase2 we want to cater to performance sensitive as well as throughput oriented workloads, which is why we’ve deployed Intel CPUs for this expansion. Monash joins the eRSA and NCI Nodes in offering this high-end capability. More on the composition of R@CMon phase 2 in the coming weeks!